diff --git a/README.md b/README.md
index 8a6bdc43a..cb8808221 100644
--- a/README.md
+++ b/README.md
@@ -2,180 +2,14 @@
A structural variation discovery pipeline for Illumina short-read whole-genome sequencing (WGS) data.
-## Table of Contents
-* [Requirements](#requirements)
-* [Citation](#citation)
-* [Acknowledgements](#acknowledgements)
-* [Quickstart](#quickstart)
-* [Pipeline Overview](#overview)
- * [Cohort mode](#cohort-mode)
- * [Single-sample mode](#single-sample-mode)
- * [gCNV model](#gcnv-training-overview)
- * [Generating a reference panel](#reference-panel-generation)
-* [Module Descriptions](#descriptions)
- * [GatherSampleEvidence](#gather-sample-evidence) - Raw callers and evidence collection
- * [EvidenceQC](#evidence-qc) - Batch QC
- * [TrainGCNV](#gcnv-training) - gCNV model creation
- * [GatherBatchEvidence](#gather-batch-evidence) - Batch evidence merging, BAF generation, and depth callers
- * [ClusterBatch](#cluster-batch) - Site clustering
- * [GenerateBatchMetrics](#generate-batch-metrics) - Site metrics
- * [FilterBatch](#filter-batch) - Filtering
- * [MergeBatchSites](#merge-batch-sites) - Cross-batch site merging
- * [GenotypeBatch](#genotype-batch) - Genotyping
- * [RegenotypeCNVs](#regenotype-cnvs) - Genotype refinement (optional)
- * [MakeCohortVcf](#make-cohort-vcf) - Cross-batch integration, complex event resolution, and VCF cleanup
- * [JoinRawCalls](#join-raw-calls) - Merges unfiltered calls across batches
- * [SVConcordance](#svconcordance) - Calculates genotype concordance with raw calls
- * [FilterGenotypes](#filter-genotypes) - Performs genotype filtering
- * [AnnotateVcf](#annotate-vcf) - Functional and allele frequency annotation
- * [Module 09](#module09) - QC and Visualization
- * Additional modules - Mosaic and de novo
-* [CI/CD](#cicd)
-* [Troubleshooting](#troubleshooting)
+For technical documentation on GATK-SV, including how to run the pipeline, please refer to our website.
-
-## Requirements
-
-
-### Deployment and execution:
-* A [Google Cloud](https://cloud.google.com/) account.
-* A workflow execution system supporting the [Workflow Description Language](https://openwdl.org/) (WDL), either:
- * [Cromwell](https://github.com/broadinstitute/cromwell) (v36 or higher). A dedicated server is highly recommended.
- * or [Terra](https://terra.bio/) (note preconfigured GATK-SV workflows are not yet available for this platform)
-* Recommended: [cromshell](https://github.com/broadinstitute/cromshell) for interacting with a dedicated Cromwell server.
-* Recommended: [WOMtool](https://cromwell.readthedocs.io/en/stable/WOMtool/) for validating WDL/json files.
-
-#### Alternative backends
-Because GATK-SV has been tested only on the Google Cloud Platform (GCP), we are unable to provide specific guidance or support for other execution platforms including HPC clusters and AWS. Contributions from the community to improve portability between backends will be considered on a case-by-case-basis. We ask contributors to please adhere to the following guidelines when submitting issues and pull requests:
-
-1. Code changes must be functionally equivalent on GCP backends, i.e. not result in changed output
-2. Increases to cost and runtime on GCP backends should be minimal
-3. Avoid adding new inputs and tasks to workflows. Simpler changes are more likely to be approved, e.g. small in-line changes to scripts or WDL task command sections
-4. Avoid introducing new code paths, e.g. conditional statements
-5. Additional backend-specific scripts, workflows, tests, and Dockerfiles will not be approved
-6. Changes to Dockerfiles may require extensive testing before approval
-
-We still encourage members of the community to adapt GATK-SV for non-GCP backends and share code on forked repositories. Here are a some considerations:
-* Refer to Cromwell's [documentation](https://cromwell.readthedocs.io/en/stable/backends/Backends/) for configuration instructions.
-* The handling and ordering of `glob` commands may differ between platforms.
-* Shell commands that are potentially destructive to input files (e.g. `rm`, `mv`, `tabix`) can cause unexpected behavior on shared filesystems. Enabling [copy localization](https://cromwell.readthedocs.io/en/stable/Configuring/#local-filesystem-options) may help to more closely replicate the behavior on GCP.
-* For clusters that do not support Docker, Singularity is an alternative. See [Cromwell documentation on Singularity](https://cromwell.readthedocs.io/en/stable/tutorials/Containers/#singularity).
-* The GATK-SV pipeline takes advantage of the massive parallelization possible in the cloud. Local backends may not have the resources to execute all of the workflows. Workflows that use fewer resources or that are less parallelized may be more successful. For instance, some users have been able to run [GatherSampleEvidence](#gather-sample-evidence) on a SLURM cluster.
-
-### Data:
-* Illumina short-read whole-genome CRAMs or BAMs, aligned to hg38 with [bwa-mem](https://github.com/lh3/bwa). BAMs must also be indexed.
-* Family structure definitions file in [PED format](#ped-format).
-
-#### PED file format
-The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
-* The file must be tab-delimited.
-* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](#evidence-qc)) should be entered as sex = 0.
-* All family, individual, and parental IDs must conform to the [sample ID requirements](#sampleids).
-* Missing parental IDs should be entered as 0.
-* Header lines are allowed if they begin with a # character.
-To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
-
-#### Sample Exclusion
-We recommend filtering out samples with a high percentage of improperly paired reads (>10% or an outlier for your data) as technical outliers prior to running [GatherSampleEvidence](#gather-sample-evidence). A high percentage of improperly paired reads may indicate issues with library prep, degradation, or contamination. Artifactual improperly paired reads could cause incorrect SV calls, and these samples have been observed to have longer runtimes and higher compute costs for [GatherSampleEvidence](#gather-sample-evidence).
-
-#### Sample ID requirements:
-
-Sample IDs must:
-* Be unique within the cohort
-* Contain only alphanumeric characters and underscores (no dashes, whitespace, or special characters)
-
-Sample IDs should not:
-* Contain only numeric characters
-* Be a substring of another sample ID in the same cohort
-* Contain any of the following substrings: `chr`, `name`, `DEL`, `DUP`, `CPX`, `CHROM`
-
-The same requirements apply to family IDs in the PED file, as well as batch IDs and the cohort ID provided as workflow inputs.
-
-Sample IDs are provided to [GatherSampleEvidence](#gather-sample-evidence) directly and need not match sample names from the BAM/CRAM headers. `GetSampleID.wdl` can be used to fetch BAM sample IDs and also generates a set of alternate IDs that are considered safe for this pipeline; alternatively, [this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py) transforms a list of sample IDs to fit these requirements. Currently, sample IDs can be replaced again in [GatherBatchEvidence](#gather-batch-evidence) - to do so, set the parameter `rename_samples = True` and provide updated sample IDs via the `samples` parameter.
-
-The following inputs will need to be updated with the transformed sample IDs:
-* Sample ID list for [GatherSampleEvidence](#gather-sample-evidence) or [GatherBatchEvidence](#gather-batch-evidence)
-* PED file
-
-
-## Citation
-Please cite the following publication:
-[Collins, Brand, et al. 2020. "A structural variation reference for medical and population genetics." Nature 581, 444-451.](https://doi.org/10.1038/s41586-020-2287-8)
-
-Additional references:
-[Werling et al. 2018. "An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder." Nature genetics 50.5, 727-736.](http://dx.doi.org/10.1038/s41588-018-0107-y)
-
-
-## Acknowledgements
-The following resources were produced using data from the All of Us Research Program and have been approved by the Program for public dissemination:
-
-* Genotype filtering model: "aou_recalibrate_gq_model_file" in "inputs/values/resources_hg38.json"
-
-The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276. In addition, the All of Us Research Program would not be possible without the partnership of its participants.
-
-
-## Quickstart
-
-#### WDLs
-There are two scripts for running the full pipeline:
-* `wdl/GATKSVPipelineBatch.wdl`: Runs GATK-SV on a batch of samples.
-* `wdl/GATKSVPipelineSingleSample.wdl`: Runs GATK-SV on a single sample, given a reference panel
-
-#### Building inputs
-Example workflow inputs can be found in `/inputs`. Build using `scripts/inputs/build_default_inputs.sh`, which
-generates input jsons in `/inputs/build`. All required resources are available in public
-Google buckets.
-
-#### MELT
-**Important**: MELT has been replaced with [Scramble](https://github.com/GeneDx/scramble) for mobile element calling. While it is still possible to run GATK-SV with MELT, we no longer support it as a caller. It will be fully deprecated in the future.
-
-Due to licensing restrictions, we cannot redistribute MELT binaries or input files, including the docker image. Some default input files contain MELT inputs that are NOT public (see [Requirements](#requirements)) including:
-
-* `GATKSVPipelineSingleSample.melt_docker` and `GATKSVPipelineBatch.melt_docker` - MELT docker URI (see [Docker readme](https://github.com/talkowski-lab/gatk-sv-v1/blob/master/dockerfiles/README.md))
-* `GATKSVPipelineSingleSample.ref_std_melt_vcfs` - Standardized MELT VCFs ([GatherBatchEvidence](#gather-batch-evidence))
-
-The input values are provided only as placeholders. In some workflows, MELT must be enabled with appropriate settings, by providing optional MELT inputs and/or with an explicit option e.g. `GATKSVPipelineBatch.use_melt` to `true`. We do not recommend running both Scramble and MELT together.
-
-#### Execution
-We recommend running the pipeline on a dedicated [Cromwell](https://github.com/broadinstitute/cromwell) server with a [cromshell](https://github.com/broadinstitute/cromshell) client. A batch run can be started with the following commands:
-
-```
-> mkdir gatksv_run && cd gatksv_run
-> mkdir wdl && cd wdl
-> cp $GATK_SV_ROOT/wdl/*.wdl .
-> zip dep.zip *.wdl
-> cd ..
-> bash scripts/inputs/build_default_inputs.sh -d $GATK_SV_ROOT
-> cp $GATK_SV_ROOT/inputs/build/ref_panel_1kg/test/GATKSVPipelineBatch/GATKSVPipelineBatch.json GATKSVPipelineBatch.my_run.json
-> cromshell submit wdl/GATKSVPipelineBatch.wdl GATKSVPipelineBatch.my_run.json cromwell_config.json wdl/dep.zip
-```
-
-where `cromwell_config.json` is a Cromwell [workflow options file](https://cromwell.readthedocs.io/en/stable/wf_options/Overview/). Note users will need to re-populate batch/sample-specific parameters (e.g. BAMs and sample IDs).
-
-## Pipeline Overview
-The pipeline consists of a series of modules that perform the following:
-* [GatherSampleEvidence](#gather-sample-evidence): SV evidence collection, including calls from a configurable set of algorithms (Manta, Scramble, and Wham), read depth (RD), split read positions (SR), and discordant pair positions (PE).
-* [EvidenceQC](#evidence-qc): Dosage bias scoring and ploidy estimation
-* [GatherBatchEvidence](#gather-batch-evidence): Copy number variant calling using cn.MOPS and GATK gCNV; B-allele frequency (BAF) generation; call and evidence aggregation
-* [ClusterBatch](#cluster-batch): Variant clustering
-* [GenerateBatchMetrics](#generate-batch-metrics): Variant filtering metric generation
-* [FilterBatch](#filter-batch): Variant filtering; outlier exclusion
-* [GenotypeBatch](#genotype-batch): Genotyping
-* [MakeCohortVcf](#make-cohort-vcf): Cross-batch integration; complex variant resolution and re-genotyping; vcf cleanup
-* [JoinRawCalls](#join-raw-calls): Merges unfiltered calls across batches
-* [SVConcordance](#svconcordance): Calculates genotype concordance with raw calls
-* [FilterGenotypes](#filter-genotypes): Performs genotype filtering
-* [AnnotateVcf](#annotate-vcf): Annotations, including functional annotation, allele frequency (AF) annotation and AF annotation with external population callsets
-* [Module 09](#module09): Visualization, including scripts that generates IGV screenshots and rd plots.
-* Additional modules to be added: de novo and mosaic scripts
-
-
-Repository structure:
+## Repository structure
+* `/carrot`: [Carrot](https://github.com/broadinstitute/carrot) tests
* `/dockerfiles`: Resources for building pipeline docker images
* `/inputs`: files for generating workflow inputs
* `/templates`: Input json file templates
* `/values`: Input values used to populate templates
-* `/wdl`: WDLs running the pipeline. There is a master WDL for running each module, e.g. `ClusterBatch.wdl`.
* `/scripts`: scripts for running tests, building dockers, and analyzing cromwell metadata files
* `/src`: main pipeline scripts
* `/RdTest`: scripts for depth testing
@@ -183,435 +17,9 @@ Repository structure:
* `/svqc`: Python module for checking that pipeline metrics fall within acceptable limits
* `/svtest`: Python module for generating various summary metrics from module outputs
* `/svtk`: Python module of tools for SV-related datafile parsing and analysis
- * `/WGD`: whole-genome dosage scoring scripts
-
-
-## Cohort mode
-A minimum cohort size of 100 is required, and a roughly equal number of males and females is recommended. For modest cohorts (~100-500 samples), the pipeline can be run as a single batch using `GATKSVPipelineBatch.wdl`.
-
-For larger cohorts, samples should be split up into batches of about 100-500 samples. Refer to the [Batching](#batching) section for further guidance on creating batches.
-
-The pipeline should be executed as follows:
-* Modules [GatherSampleEvidence](#gather-sample-evidence) and [EvidenceQC](#evidence-qc) can be run on arbitrary cohort partitions
-* Modules [GatherBatchEvidence](#gather-batch-evidence), [ClusterBatch](#cluster-batch), [GenerateBatchMetrics](#generate-batch-metrics), and [FilterBatch](#filter-batch) are run separately per batch
-* [GenotypeBatch](#genotype-batch) is run separately per batch, using filtered variants ([FilterBatch](#filter-batch) output) combined across all batches
-* [MakeCohortVcf](#make-cohort-vcf) and beyond are run on all batches together
-
-Note: [GatherBatchEvidence](#gather-batch-evidence) requires a [trained gCNV model](#gcnv-training).
-
-#### Batching
-For larger cohorts, samples should be split up into batches of about 100-500 samples with similar characteristics. We recommend batching based on overall coverage and dosage score (WGD), which can be generated in [EvidenceQC](#evidence-qc). An example batching process is outlined below:
-1. Divide the cohort into PCR+ and PCR- samples
-2. Partition the samples by median coverage from [EvidenceQC](#evidence-qc), grouping samples with similar median coverage together. The end goal is to divide the cohort into roughly equal-sized batches of about 100-500 samples; if your partitions based on coverage are larger or uneven, you can partition the cohort further in the next step to obtain the final batches.
-3. Optionally, divide the samples further by dosage score (WGD) from [EvidenceQC](#evidence-qc), grouping samples with similar WGD score together, to obtain roughly equal-sized batches of about 100-500 samples
-4. Maintain a roughly equal sex balance within each batch, based on sex assignments from [EvidenceQC](#evidence-qc)
-
-
-## Single-sample mode
-`GATKSVPipelineSingleSample.wdl` runs the pipeline on a single sample using a fixed reference panel. An example run with reference panel containing 156 samples from the [NYGC 1000G Terra workspace](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019) can be found in `inputs/build/NA12878/test` after [building inputs](#building-inputs)).
-
-## gCNV Training
-Both the cohort and single-sample modes use the [GATK-gCNV](https://gatk.broadinstitute.org/hc/en-us/articles/360035531152) depth calling pipeline, which requires a [trained model](#gcnv-training) as input. The samples used for training should be technically homogeneous and similar to the samples to be processed (i.e. same sample type, library prep protocol, sequencer, sequencing center, etc.). The samples to be processed may comprise all or a subset of the training set. For small, relatively homogenous cohorts, a single gCNV model is usually sufficient. If a cohort contains multiple data sources, we recommend training a separate model for each [batch](#batching) or group of batches with similar dosage score (WGD). The model may be trained on all or a subset of the samples to which it will be applied; a reasonable default is 100 randomly-selected samples from the batch (the random selection can be done as part of the workflow by specifying a number of samples to the `n_samples_subsample` input parameter in `/wdl/TrainGCNV.wdl`).
-
-## Generating a reference panel
-New reference panels can be generated easily from a single run of the `GATKSVPipelineBatch` workflow. If using a Cromwell server, we recommend copying the outputs to a permanent location by adding the following option to the workflow configuration file:
-```
- "final_workflow_outputs_dir" : "gs://my-outputs-bucket",
- "use_relative_output_paths": false,
-```
-Here is an example of how to generate workflow input jsons from `GATKSVPipelineBatch` workflow metadata:
-```
-> cromshell -t60 metadata 38c65ca4-2a07-4805-86b6-214696075fef > metadata.json
-> python scripts/inputs/create_test_batch.py \
- --execution-bucket gs://my-exec-bucket \
- --final-workflow-outputs-dir gs://my-outputs-bucket \
- metadata.json \
- > inputs/values/my_ref_panel.json
-> # Build test files for batched workflows
-> python scripts/inputs/build_inputs.py \
- inputs/values \
- inputs/templates/test \
- inputs/build/my_ref_panel/test \
- -a '{ "test_batch" : "ref_panel_1kg" }'
-> # Build test files for the single-sample workflow
-> python scripts/inputs/build_inputs.py \
- inputs/values \
- inputs/templates/test/GATKSVPipelineSingleSample \
- inputs/build/NA19240/test_my_ref_panel \
- -a '{ "single_sample" : "test_single_sample_NA19240", "ref_panel" : "my_ref_panel" }'
-> # Build files for a Terra workspace
-> python scripts/inputs/build_inputs.py \
- inputs/values \
- inputs/templates/terra_workspaces/single_sample \
- inputs/build/NA12878/terra_my_ref_panel \
- -a '{ "single_sample" : "test_single_sample_NA12878", "ref_panel" : "my_ref_panel" }'
-```
-Note that the inputs to `GATKSVPipelineBatch` may be used as resources for the reference panel and therefore should also be in a permanent location.
-
-## Module Descriptions
-The following sections briefly describe each module and highlights inter-dependent input/output files. Note that input/output mappings can also be gleaned from `GATKSVPipelineBatch.wdl`, and example input templates for each module can be found in `/inputs/templates/test`.
-
-## GatherSampleEvidence
-*Formerly Module00a*
-
-Runs raw evidence collection on each sample with the following SV callers: [Manta](https://github.com/Illumina/manta), [Wham](https://github.com/zeeev/wham), [Scramble](https://github.com/GeneDx/scramble), and/or [MELT](https://melt.igs.umaryland.edu/). For guidance on pre-filtering prior to `GatherSampleEvidence`, refer to the [Sample Exclusion](#sample-exclusion) section.
-
-The `scramble_clusters` and `scramble_table` are generated as outputs for troubleshooting purposes but not consumed by any downstream workflows.
-
-Note: a list of sample IDs must be provided. Refer to the [sample ID requirements](#sampleids) for specifications of allowable sample IDs. IDs that do not meet these requirements may cause errors.
-
-#### Inputs:
-* Per-sample BAM or CRAM files aligned to hg38. Index files (`.bai`) must be provided if using BAMs.
-
-#### Outputs:
-* Caller VCFs (Manta, Scramble, MELT, and/or Wham)
-* Binned read counts file
-* Split reads (SR) file
-* Discordant read pairs (PE) file
-* Scramble intermediate clusters file and table (not needed downstream)
-
-## EvidenceQC
-*Formerly Module00b*
-
-Runs ploidy estimation, dosage scoring, and optionally VCF QC. The results from this module can be used for QC and batching.
-
-For large cohorts, this workflow can be run on arbitrary cohort partitions of up to about 500 samples. Afterwards, we recommend using the results to divide samples into smaller batches (~100-500 samples) with ~1:1 male:female ratio. Refer to the [Batching](#batching) section for further guidance on creating batches.
-
-We also recommend using sex assignments generated from the ploidy estimates and incorporating them into the PED file, with sex = 0 for sex aneuploidies.
-
-#### Prerequisites:
-* [GatherSampleEvidence](#gather-sample-evidence)
-
-#### Inputs:
-* Read count files ([GatherSampleEvidence](#gather-sample-evidence))
-* (Optional) SV call VCFs ([GatherSampleEvidence](#gather-sample-evidence))
-
-#### Outputs:
-* Per-sample dosage scores with plots
-* Median coverage per sample
-* Ploidy estimates, sex assignments, with plots
-* (Optional) Outlier samples detected by call counts
-
-#### Preliminary Sample QC
-The purpose of sample filtering at this stage after EvidenceQC is to prevent very poor quality samples from interfering with the results for the rest of the callset. In general, samples that are borderline are okay to leave in, but you should choose filtering thresholds to suit the needs of your cohort and study. There will be future opportunities (as part of [FilterBatch](#filter-batch)) for filtering before the joint genotyping stage if necessary. Here are a few of the basic QC checks that we recommend:
-* Look at the X and Y ploidy plots, and check that sex assignments match your expectations. If there are discrepancies, check for sample swaps and update your PED file before proceeding.
-* Look at the dosage score (WGD) distribution and check that it is centered around 0 (the distribution of WGD for PCR- samples is expected to be slightly lower than 0, and the distribution of WGD for PCR+ samples is expected to be slightly greater than 0. Refer to the [gnomAD-SV paper](https://doi.org/10.1038/s41586-020-2287-8) for more information on WGD score). Optionally filter outliers.
-* Look at the low outliers for each SV caller (samples with much lower than typical numbers of SV calls per contig for each caller). An empty low outlier file means there were no outliers below the median and no filtering is necessary. Check that no samples had zero calls.
-* Look at the high outliers for each SV caller and optionally filter outliers; samples with many more SV calls than average may be poor quality.
-* Remove samples with autosomal aneuploidies based on the per-batch binned coverage plots of each chromosome.
-
-
-## TrainGCNV
-Trains a [gCNV](https://gatk.broadinstitute.org/hc/en-us/articles/360035531152) model for use in [GatherBatchEvidence](#gather-batch-evidence). The WDL can be found at `/wdl/TrainGCNV.wdl`. See the [gCNV training overview](#gcnv-training-overview) for more information.
-
-#### Prerequisites:
-* [GatherSampleEvidence](#gather-sample-evidence)
-* (Recommended) [EvidenceQC](#evidence-qc)
-
-#### Inputs:
-* Read count files ([GatherSampleEvidence](#gather-sample-evidence))
-
-#### Outputs:
-* Contig ploidy model tarball
-* gCNV model tarballs
-
-
-## GatherBatchEvidence
-*Formerly Module00c*
-
-Runs CNV callers ([cn.MOPS](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3351174/), [GATK-gCNV](https://gatk.broadinstitute.org/hc/en-us/articles/360035531152)) and combines single-sample raw evidence into a batch. See [above](#cohort-mode) for more information on batching.
-
-#### Prerequisites:
-* [GatherSampleEvidence](#gather-sample-evidence)
-* (Recommended) [EvidenceQC](#evidence-qc)
-* [gCNV training](#gcnv-training)
-
-#### Inputs:
-* PED file (updated with [EvidenceQC](#evidence-qc) sex assignments, including sex = 0 for sex aneuploidies. Calls will not be made on sex chromosomes when sex = 0 in order to avoid generating many confusing calls or upsetting normalized copy numbers for the batch.)
-* Read count, BAF, PE, SD, and SR files ([GatherSampleEvidence](#gather-sample-evidence))
-* Caller VCFs ([GatherSampleEvidence](#gather-sample-evidence))
-* Contig ploidy model and gCNV model files ([gCNV training](#gcnv-training))
-
-#### Outputs:
-* Combined read count matrix, SR, PE, and BAF files
-* Standardized call VCFs
-* Depth-only (DEL/DUP) calls
-* Per-sample median coverage estimates
-* (Optional) Evidence QC plots
-
-
-## ClusterBatch
-*Formerly Module01*
-
-Clusters SV calls across a batch.
-
-#### Prerequisites:
-* [GatherBatchEvidence](#gather-batch-evidence)
-
-#### Inputs:
-* Standardized call VCFs ([GatherBatchEvidence](#gather-batch-evidence))
-* Depth-only (DEL/DUP) calls ([GatherBatchEvidence](#gather-batch-evidence))
-
-#### Outputs:
-* Clustered SV VCFs
-* Clustered depth-only call VCF
-
-
-## GenerateBatchMetrics
-*Formerly Module02*
-
-Generates variant metrics for filtering.
-
-#### Prerequisites:
-* [ClusterBatch](#cluster-batch)
-
-#### Inputs:
-* Combined read count matrix, SR, PE, and BAF files ([GatherBatchEvidence](#gather-batch-evidence))
-* Per-sample median coverage estimates ([GatherBatchEvidence](#gather-batch-evidence))
-* Clustered SV VCFs ([ClusterBatch](#cluster-batch))
-* Clustered depth-only call VCF ([ClusterBatch](#cluster-batch))
-
-#### Outputs:
-* Metrics file
-
-
-## FilterBatch
-*Formerly Module03*
-
-Filters poor quality variants and filters outlier samples. This workflow can be run all at once with the WDL at `wdl/FilterBatch.wdl`, or it can be run in two steps to enable tuning of outlier filtration cutoffs. The two subworkflows are:
-1. FilterBatchSites: Per-batch variant filtration. Visualize SV counts per sample per type to help choose an IQR cutoff for outlier filtering, and preview outlier samples for a given cutoff
-2. FilterBatchSamples: Per-batch outlier sample filtration; provide an appropriate `outlier_cutoff_nIQR` based on the SV count plots and outlier previews from step 1. Note that not removing high outliers can result in increased compute cost and a higher false positive rate in later steps.
-
-#### Prerequisites:
-* [GenerateBatchMetrics](#generate-batch-metrics)
-
-#### Inputs:
-* Batch PED file
-* Metrics file ([GenerateBatchMetrics](#generate-batch-metrics))
-* Clustered SV and depth-only call VCFs ([ClusterBatch](#cluster-batch))
-
-#### Outputs:
-* Filtered SV (non-depth-only a.k.a. "PESR") VCF with outlier samples excluded
-* Filtered depth-only call VCF with outlier samples excluded
-* Random forest cutoffs file
-* PED file with outlier samples excluded
-
-
-## MergeBatchSites
-*Formerly MergeCohortVcfs*
-
-Combines filtered variants across batches. The WDL can be found at: `/wdl/MergeBatchSites.wdl`.
-
-#### Prerequisites:
-* [FilterBatch](#filter-batch)
-
-#### Inputs:
-* List of filtered PESR VCFs ([FilterBatch](#filter-batch))
-* List of filtered depth VCFs ([FilterBatch](#filter-batch))
-
-#### Outputs:
-* Combined cohort PESR and depth VCFs
-
-
-## GenotypeBatch
-*Formerly Module04*
-
-Genotypes a batch of samples across unfiltered variants combined across all batches.
-
-#### Prerequisites:
-* [FilterBatch](#filter-batch)
-* [MergeBatchSites](#merge-batch-sites)
-
-#### Inputs:
-* Batch PESR and depth VCFs ([FilterBatch](#filter-batch))
-* Cohort PESR and depth VCFs ([MergeBatchSites](#merge-batch-sites))
-* Batch read count, PE, and SR files ([GatherBatchEvidence](#gather-batch-evidence))
-
-#### Outputs:
-* Filtered SV (non-depth-only a.k.a. "PESR") VCF with outlier samples excluded
-* Filtered depth-only call VCF with outlier samples excluded
-* PED file with outlier samples excluded
-* List of SR pass variants
-* List of SR fail variants
-* (Optional) Depth re-genotyping intervals list
-
-
-## RegenotypeCNVs
-*Formerly Module04b*
-
-Re-genotypes probable mosaic variants across multiple batches.
-
-#### Prerequisites:
-* [GenotypeBatch](#genotype-batch)
-
-#### Inputs:
-* Per-sample median coverage estimates ([GatherBatchEvidence](#gather-batch-evidence))
-* Pre-genotyping depth VCFs ([FilterBatch](#filter-batch))
-* Batch PED files ([FilterBatch](#filter-batch))
-* Cohort depth VCF ([MergeBatchSites](#merge-batch-sites))
-* Genotyped depth VCFs ([GenotypeBatch](#genotype-batch))
-* Genotyped depth RD cutoffs file ([GenotypeBatch](#genotype-batch))
-
-#### Outputs:
-* Re-genotyped depth VCFs
-
-
-## MakeCohortVcf
-*Formerly Module0506*
-
-Combines variants across multiple batches, resolves complex variants, re-genotypes, and performs final VCF clean-up.
-
-#### Prerequisites:
-* [GenotypeBatch](#genotype-batch)
-* (Optional) [RegenotypeCNVs](#regenotype-cnvs)
-
-#### Inputs:
-* RD, PE and SR file URIs ([GatherBatchEvidence](#gather-batch-evidence))
-* Batch filtered PED file URIs ([FilterBatch](#filter-batch))
-* Genotyped PESR VCF URIs ([GenotypeBatch](#genotype-batch))
-* Genotyped depth VCF URIs ([GenotypeBatch](#genotype-batch) or [RegenotypeCNVs](#regenotype-cnvs))
-* SR pass variant file URIs ([GenotypeBatch](#genotype-batch))
-* SR fail variant file URIs ([GenotypeBatch](#genotype-batch))
-* Genotyping cutoff file URIs ([GenotypeBatch](#genotype-batch))
-* Batch IDs
-* Sample ID list URIs
-
-#### Outputs:
-* Finalized "cleaned" VCF and QC plots
-
-## JoinRawCalls
-
-Merges raw unfiltered calls across batches. Concordance between these genotypes and the joint call set usually can be indicative of variant quality and is used downstream for genotype filtering.
-
-#### Prerequisites:
-* [ClusterBatch](#cluster-batch)
-
-#### Inputs:
-* Clustered Manta, Wham, depth, Scramble, and/or MELT VCF URIs ([ClusterBatch](#cluster-batch))
-* PED file
-* Reference sequence
-
-#### Outputs:
-* VCF of clustered raw calls
-* Ploidy table
-
-## SVConcordance
-
-Computes genotype concordance metrics between all variants in the joint call set and raw calls.
-
-#### Prerequisites:
-* [MakeCohortVcf](#make-cohort-vcf)
-* [JoinRawCalls](#join-raw-calls)
-
-#### Inputs:
-* Cleaned ("eval") VCF URI ([MakeCohortVcf](#make-cohort-vcf))
-* Joined raw call ("truth") VCF URI ([JoinRawCalls](#join-raw-calls))
-* Reference dictionary URI
-
-#### Outputs:
-* VCF with concordance annotations
-
-## FilterGenotypes
-
-Performs genotype quality recalibration using a machine learning model based on [xgboost](https://github.com/dmlc/xgboost), and filters genotypes.
-
-The ML model uses the following features:
-
-* Genotype properties:
- * Non-reference and no-call allele counts
- * Genotype quality (GQ)
- * Supporting evidence types (EV) and respective genotype qualities (PE_GQ, SR_GQ, RD_GQ)
- * Raw call concordance (CONC_ST)
-* Variant properties:
- * Variant type (SVTYPE) and size (SVLEN)
- * FILTER status
- * Calling algorithms (ALGORITHMS)
- * Supporting evidence types (EVIDENCE)
- * Two-sided SR support flag (BOTHSIDES_SUPPORT)
- * Evidence overdispersion flag (PESR_GT_OVERDISPERSION)
- * SR noise flag (HIGH_SR_BACKGROUND)
- * Raw call concordance (STATUS, NON_REF_GENOTYPE_CONCORDANCE, VAR_PPV, VAR_SENSITIVITY, TRUTH_AF)
-* Reference context with respect to UCSC Genome Browser tracks:
- * RepeatMasker
- * Segmental duplications
- * Simple repeats
- * K-mer mappability (umap_s100 and umap_s24)
-
-For ease of use, we provide a model pre-trained on high-quality data with truth data derived from long-read calls:
-```
-gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gatk-sv-recalibrator.aou_phase_1.v1.model
-```
-See the SV "Genotype Filter" section on page 34 of the [All of Us Genomic Quality Report C2022Q4R9 CDR v7](https://support.researchallofus.org/hc/en-us/articles/4617899955092-All-of-Us-Genomic-Quality-Report-ARCHIVED-C2022Q4R9-CDR-v7) for further details on model training.
-
-All valid genotypes are annotated with a "scaled logit" (SL) score, which is rescaled to non-negative adjusted GQs on [1, 99]. Note that the rescaled GQs should *not* be interpreted as probabilities. Original genotype qualities are retained in the OGQ field.
-
-A more positive SL score indicates higher probability that the given genotype is not homozygous for the reference allele. Genotypes are therefore filtered using SL thresholds that depend on SV type and size. This workflow also generates QC plots using the [MainVcfQc](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MainVcfQc.wdl) workflow to review call set quality (see below for recommended practices).
-
-This workflow can be run in one of two modes:
-
-1. (Recommended) The user explicitly provides a set of SL cutoffs through the `sl_filter_args` parameter, e.g.
- ```
- "--small-del-threshold 93 --medium-del-threshold 150 --small-dup-threshold -51 --medium-dup-threshold -4 --ins-threshold -13 --inv-threshold -19"
- ```
- Genotypes with SL scores less than the cutoffs are set to no-call (`./.`). The above values were taken directly from Appendix N of the [All of Us Genomic Quality Report C2022Q4R9 CDR v7 ](https://support.researchallofus.org/hc/en-us/articles/4617899955092-All-of-Us-Genomic-Quality-Report-ARCHIVED-C2022Q4R9-CDR-v7). Users should adjust the thresholds depending on data quality and desired accuracy. Please see the arguments in [this script](https://github.com/broadinstitute/gatk-sv/blob/main/src/sv-pipeline/scripts/apply_sl_filter.py) for all available options.
-
-2. (Advanced) The user provides truth labels for a subset of non-reference calls, and SL cutoffs are automatically optimized. These truth labels should be provided as a json file in the following format:
- ```
- {
- "sample_1": {
- "good_variant_ids": ["variant_1", "variant_3"],
- "bad_variant_ids": ["variant_5", "variant_10"]
- },
- "sample_2": {
- "good_variant_ids": ["variant_2", "variant_13"],
- "bad_variant_ids": ["variant_8", "variant_11"]
- }
- }
- ```
- where "good_variant_ids" and "bad_variant_ids" are lists of variant IDs corresponding to non-reference (i.e. het or hom-var) sample genotypes that are true positives and false positives, respectively. SL cutoffs are optimized by maximizing the [F-score](https://en.wikipedia.org/wiki/F-score) with "beta" parameter `fmax_beta`, which modulates the weight given to precision over recall (lower values give higher precision).
-
-In both modes, the workflow additionally filters variants based on the "no-call rate", the proportion of genotypes that were filtered in a given variant. Variants exceeding the `no_call_rate_cutoff` are assigned a `HIGH_NCR` filter status.
-
-We recommend users observe the following basic criteria to assess the overall quality of the filtered call set:
-
-* Number of PASS variants (excluding BND) between 7,000 and 11,000.
-* At least 75% of variants in Hardy-Weinberg equilibrium (HWE). Note that this could be lower, depending on how how closely the cohort adheres to the assumptions of the Hardy-Weinberg model. However, HWE is expected to at least improve after filtering.
-* Low *de novo* inheritance rate (if applicable), typically 5-10%.
-
-These criteria can be assessed from the plots in the `main_vcf_qc_tarball` output, which is generated by default.
-
-#### Prerequisites:
-* [SVConcordance](#svconcordance)
-
-#### Inputs:
-* VCF with genotype concordance annotations URI ([SVConcordance](#svconcordance))
-* Ploidy table URI ([JoinRawCalls](#join-raw-calls))
-* GQRecalibrator model URI
-* Either a set of SL cutoffs or truth labels
-
-#### Outputs:
-* Filtered VCF
-* Call set QC plots (optional)
-* Optimized SL cutoffs with filtering QC plots and data tables (if running mode [2] with truth labels)
-* VCF with only SL annotation and GQ recalibration (before filtering)
-
-## AnnotateVcf
-*Formerly Module08Annotation*
-
-Add annotations, such as the inferred function and allele frequencies of variants, to final VCF.
-
-Annotations methods include:
-* Functional annotation - The GATK tool [SVAnnotate](https://gatk.broadinstitute.org/hc/en-us/articles/13832752531355-SVAnnotate) is used to annotate SVs with inferred functional consequence on protein-coding regions, regulatory regions such as UTR and promoters, and other non-coding elements.
-* Allele Frequency annotation - annotate SVs with their allele frequencies across all samples, and samples of specific sex, as well as specific sub-populations.
-* Allele Frequency annotation with external callset - annotate SVs with the allele frequencies of their overlapping SVs in another callset, eg. gnomad SV callset.
-
-## Module 09 (in development)
-Visualize SVs with [IGV](http://software.broadinstitute.org/software/igv/) screenshots and read depth plots.
-
-Visualization methods include:
-* RD Visualization - generate RD plots across all samples, ideal for visualizing large CNVs.
-* IGV Visualization - generate IGV plots of each SV for individual sample, ideal for visualizing de novo small SVs.
-* Module09.visualize.wdl - generate RD plots and IGV plots, and combine them for easy review.
+ * `/WGD`: whole-genome dosage score scripts
+* `/wdl`: WDLs running the pipeline. There is a master WDL for running each module, e.g. `ClusterBatch.wdl`.
+* `/website`: website code
## CI/CD
This repository is maintained following the norms of
@@ -620,25 +28,3 @@ GATK-SV CI/CD is developed as a set of Github Actions
workflows that are available under the `.github/workflows`
directory. Please refer to the [workflow's README](.github/workflows/README.md)
for their current coverage and setup.
-
-## Troubleshooting
-
-### VM runs out of memory or disk
-* Default pipeline settings are tuned for batches of 100 samples. Larger batches or cohorts may require additional VM resources. Most runtime attributes can be modified through the `RuntimeAttr` inputs. These are formatted like this in the json:
-```
-"MyWorkflow.runtime_attr_override": {
- "disk_gb": 100,
- "mem_gb": 16
-},
-```
-Note that a subset of the struct attributes can be specified. See `wdl/Structs.wdl` for available attributes.
-
-
-### Calculated read length causes error in MELT workflow
-
-Example error message from `GatherSampleEvidence.MELT.GetWgsMetrics`:
-```
-Exception in thread "main" java.lang.ArrayIndexOutOfBoundsException: The requested index 701766 is out of counter bounds. Possible cause of exception can be wrong READ_LENGTH parameter (much smaller than actual read length)
-```
-
-This error message was observed for a sample with an average read length of 117, but for which half the reads were of length 90 and half were of length 151. As a workaround, override the calculated read length by providing a `read_length` input of 151 (or the expected read length for the sample in question) to `GatherSampleEvidence`.
diff --git a/wdl/GenotypeBatch.wdl b/wdl/GenotypeBatch.wdl
index 39855a81e..04343733c 100644
--- a/wdl/GenotypeBatch.wdl
+++ b/wdl/GenotypeBatch.wdl
@@ -28,7 +28,7 @@ workflow GenotypeBatch {
File? pesr_exclude_list # Required unless skipping training
File splitfile
File? splitfile_index
- String? reference_build #hg19 or hg38, Required unless skipping training
+ String? reference_build # Must be hg38, Required unless skipping training
File bin_exclude
File ref_dict
# If all specified, training will be skipped (for single sample pipeline)
diff --git a/website/docs/acknowledgements.md b/website/docs/acknowledgements.md
new file mode 100644
index 000000000..4ee89fbdf
--- /dev/null
+++ b/website/docs/acknowledgements.md
@@ -0,0 +1,18 @@
+---
+title: Acknowledgements
+description: Acknowledgements
+sidebar_position: 10
+---
+
+The following resources were produced using data from the [All of Us Research Program](https://allofus.nih.gov/)
+and have been approved by the Program for public dissemination:
+
+* Genotype filtering model: "aou_recalibrate_gq_model_file" in "inputs/values/resources_hg38.json"
+
+The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional
+Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1
+OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers:
+HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24
+OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2
+OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276. In addition, the All
+of Us Research Program would not be possible without the partnership of its participants.
diff --git a/website/docs/advanced/_category_.json b/website/docs/advanced/_category_.json
index 99c08a85c..c88d64045 100644
--- a/website/docs/advanced/_category_.json
+++ b/website/docs/advanced/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Advanced Guides",
- "position": 8,
+ "position": 9,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/advanced/build_inputs.md b/website/docs/advanced/build_inputs.md
index 2beb1993a..66fb6cb2f 100644
--- a/website/docs/advanced/build_inputs.md
+++ b/website/docs/advanced/build_inputs.md
@@ -1,7 +1,7 @@
---
title: Building inputs
description: Building work input json files
-sidebar_position: 1
+sidebar_position: 3
slug: build_inputs
---
@@ -43,8 +43,6 @@ You may run the following commands to get these example inputs.
└── test
```
-## Building inputs for specific use-cases (Advanced)
-
### Build for batched workflows
```shell
@@ -55,69 +53,3 @@ python scripts/inputs/build_inputs.py \
-a '{ "test_batch" : "ref_panel_1kg" }'
```
-
-### Generating a reference panel
-
-This section only applies to the single-sample mode.
-New reference panels can be generated from a single run of the
-`GATKSVPipelineBatch` workflow.
-If using a Cromwell server, we recommend copying the outputs to a
-permanent location by adding the following option to the
-[workflow configuration](https://cromwell.readthedocs.io/en/latest/wf_options/Overview/)
-file:
-
-```json
-"final_workflow_outputs_dir" : "gs://my-outputs-bucket",
-"use_relative_output_paths": false,
-```
-
-Here is an example of how to generate workflow input jsons from `GATKSVPipelineBatch` workflow metadata:
-
-1. Get metadata from Cromwshell.
-
- ```shell
- cromshell -t60 metadata 38c65ca4-2a07-4805-86b6-214696075fef > metadata.json
- ```
-
-2. Run the script.
-
- ```shell
- python scripts/inputs/create_test_batch.py \
- --execution-bucket gs://my-exec-bucket \
- --final-workflow-outputs-dir gs://my-outputs-bucket \
- metadata.json \
- > inputs/values/my_ref_panel.json
- ```
-
-3. Build test files for batched workflows (google cloud project id required).
-
- ```shell
- python scripts/inputs/build_inputs.py \
- inputs/values \
- inputs/templates/test \
- inputs/build/my_ref_panel/test \
- -a '{ "test_batch" : "ref_panel_1kg" }'
- ```
-
-4. Build test files for the single-sample workflow
-
- ```shell
- python scripts/inputs/build_inputs.py \
- inputs/values \
- inputs/templates/test/GATKSVPipelineSingleSample \
- inputs/build/NA19240/test_my_ref_panel \
- -a '{ "single_sample" : "test_single_sample_NA19240", "ref_panel" : "my_ref_panel" }'
- ```
-
-5. Build files for a Terra workspace.
-
- ```shell
- python scripts/inputs/build_inputs.py \
- inputs/values \
- inputs/templates/terra_workspaces/single_sample \
- inputs/build/NA12878/terra_my_ref_panel \
- -a '{ "single_sample" : "test_single_sample_NA12878", "ref_panel" : "my_ref_panel" }'
- ```
-
-Note that the inputs to `GATKSVPipelineBatch` may be used as resources
-for the reference panel and therefore should also be in a permanent location.
diff --git a/website/docs/advanced/build_ref_panel.md b/website/docs/advanced/build_ref_panel.md
new file mode 100644
index 000000000..263a874fd
--- /dev/null
+++ b/website/docs/advanced/build_ref_panel.md
@@ -0,0 +1,73 @@
+---
+title: Building reference panels
+description: Building reference panels for the single-sample pipeline
+sidebar_position: 4
+slug: build_ref_panel
+---
+
+A custom reference panel for the [single-sample mode](/docs/gs/calling_modes#single-sample-mode) can be generated most easily using the
+[GATKSVPipelineBatch](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/GATKSVPipelineBatch.wdl) workflow.
+This must be run on a standalone Cromwell server, as the workflow is unstable on Terra.
+
+:::note
+Reference panels can also be generated by running the pipeline through joint calling on Terra, but there is
+currently no solution for automatically updating inputs.
+:::
+
+We recommend copying the outputs from a Cromwell run to a permanent location by adding the following option to
+the workflow configuration file:
+```
+ "final_workflow_outputs_dir" : "gs://my-outputs-bucket",
+ "use_relative_output_paths": false,
+```
+
+Here is an example of how to generate workflow input jsons from `GATKSVPipelineBatch` workflow metadata:
+
+1. Get metadata from Cromwshell.
+
+ ```shell
+ cromshell -t60 metadata 38c65ca4-2a07-4805-86b6-214696075fef > metadata.json
+ ```
+
+2. Run the script.
+
+ ```shell
+ python scripts/inputs/create_test_batch.py \
+ --execution-bucket gs://my-exec-bucket \
+ --final-workflow-outputs-dir gs://my-outputs-bucket \
+ metadata.json \
+ > inputs/values/my_ref_panel.json
+ ```
+
+3. Build test files for batched workflows (google cloud project id required).
+
+ ```shell
+ python scripts/inputs/build_inputs.py \
+ inputs/values \
+ inputs/templates/test \
+ inputs/build/my_ref_panel/test \
+ -a '{ "test_batch" : "ref_panel_1kg" }'
+ ```
+
+4. Build test files for the single-sample workflow
+
+ ```shell
+ python scripts/inputs/build_inputs.py \
+ inputs/values \
+ inputs/templates/test/GATKSVPipelineSingleSample \
+ inputs/build/NA19240/test_my_ref_panel \
+ -a '{ "single_sample" : "test_single_sample_NA19240", "ref_panel" : "my_ref_panel" }'
+ ```
+
+5. Build files for a Terra workspace.
+
+ ```shell
+ python scripts/inputs/build_inputs.py \
+ inputs/values \
+ inputs/templates/terra_workspaces/single_sample \
+ inputs/build/NA12878/terra_my_ref_panel \
+ -a '{ "single_sample" : "test_single_sample_NA12878", "ref_panel" : "my_ref_panel" }'
+ ```
+
+Note that the inputs to `GATKSVPipelineBatch` may be used as resources
+for the reference panel and therefore should also be in a permanent location.
diff --git a/website/docs/advanced/development/_category_.json b/website/docs/advanced/cromwell/_category_.json
similarity index 53%
rename from website/docs/advanced/development/_category_.json
rename to website/docs/advanced/cromwell/_category_.json
index c924cd2ab..6efde5537 100644
--- a/website/docs/advanced/development/_category_.json
+++ b/website/docs/advanced/cromwell/_category_.json
@@ -1,6 +1,6 @@
{
- "label": "Development",
- "position": 6,
+ "label": "Cromwell",
+ "position": 1,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/advanced/development/cromwell.md b/website/docs/advanced/cromwell/overview.md
similarity index 97%
rename from website/docs/advanced/development/cromwell.md
rename to website/docs/advanced/cromwell/overview.md
index f9c4ef3d2..84ff94ae4 100644
--- a/website/docs/advanced/development/cromwell.md
+++ b/website/docs/advanced/cromwell/overview.md
@@ -1,6 +1,6 @@
---
-title: Cromwell
-description: Running GATK-SV on Cromwell
+title: Overview
+description: Introduction to Cromwell
sidebar_position: 0
---
diff --git a/website/docs/gs/quick_start.md b/website/docs/advanced/cromwell/quick_start.md
similarity index 93%
rename from website/docs/gs/quick_start.md
rename to website/docs/advanced/cromwell/quick_start.md
index b225f7837..a280241bc 100644
--- a/website/docs/gs/quick_start.md
+++ b/website/docs/advanced/cromwell/quick_start.md
@@ -1,18 +1,16 @@
---
-title: Quick Start
-description: Run the pipeline on demo data.
+title: Run
+description: Running GATK-SV on Cromwell
sidebar_position: 1
slug: ./qs
---
-This page provides steps for running the pipeline using demo data.
-
# Quick Start on Cromwell
This section walks you through the steps of running pipeline using
demo data on a managed Cromwell server.
-### Setup Environment
+### Environment Setup
- A running instance of a Cromwell server.
diff --git a/website/docs/advanced/docker/_category_.json b/website/docs/advanced/docker/_category_.json
index e88fd355f..7d3dfa426 100644
--- a/website/docs/advanced/docker/_category_.json
+++ b/website/docs/advanced/docker/_category_.json
@@ -1,6 +1,6 @@
{
- "label": "Docker Images",
- "position": 7,
+ "label": "Docker builds",
+ "position": 2,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/best_practices.md b/website/docs/best_practices.md
new file mode 100644
index 000000000..4c0695d58
--- /dev/null
+++ b/website/docs/best_practices.md
@@ -0,0 +1,18 @@
+---
+title: Best Practices Guide
+description: Guide for using GATK-SV
+sidebar_position: 4
+---
+
+A comprehensive guide for the single-sample calling mode is available in [GATK Best Practices for Structural Variation
+Discovery on Single Samples](https://gatk.broadinstitute.org/hc/en-us/articles/9022653744283-GATK-Best-Practices-for-Structural-Variation-Discovery-on-Single-Samples).
+This material covers basic concepts of structural variant calling, specifics of SV VCF formatting, and
+advanced troubleshooting that also apply to the joint calling mode as well. This guide is intended to supplement
+documentation found here.
+
+Users should also review the [Getting Started](/docs/gs/overview) section before attempting to perform SV calling.
+
+The following sections also contain recommendations pertaining to data and call set QC:
+
+- Preliminary sample QC in the [EvidenceQc module](/docs/modules/eqc#preliminary-sample-qc).
+- Assessment of completed call sets can be found on the [MainVcfQc module page](/docs/modules/mvqc).
diff --git a/website/docs/run/_category_.json b/website/docs/execution/_category_.json
similarity index 73%
rename from website/docs/run/_category_.json
rename to website/docs/execution/_category_.json
index eb46f4d4c..f6b285b53 100644
--- a/website/docs/run/_category_.json
+++ b/website/docs/execution/_category_.json
@@ -1,5 +1,5 @@
{
- "label": "Run",
+ "label": "Execution",
"position": 4,
"link": {
"type": "generated-index"
diff --git a/website/docs/execution/joint.md b/website/docs/execution/joint.md
new file mode 100644
index 000000000..176dfbd07
--- /dev/null
+++ b/website/docs/execution/joint.md
@@ -0,0 +1,323 @@
+---
+title: Joint calling
+description: Run the pipeline on a cohort
+sidebar_position: 4
+slug: joint
+---
+
+## Terra workspace
+Users should clone the Terra joint calling workspace (TODO)
+which is configured with a demo sample set.
+Refer to the following sections for instructions on how to run the pipeline on your data using this workspace.
+
+### Default data
+The demonstration data in this workspace is 312 publicly-available 1000 Genomes Project samples from the
+[NYGC/AnVIL high coverage data set](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019),
+divided into two equally-sized batches.
+
+## Pipeline Expectations
+### What does it do?
+This pipeline performs structural variation discovery from CRAMs, joint genotyping, and variant resolution on a cohort
+of samples.
+
+### Required inputs
+The following inputs must be provided for each sample in the cohort, via the sample table described in **Workspace
+Setup** step 2:
+
+|Input Type|Input Name|Description|
+|---------|--------|--------------|
+|`String`|`sample_id`|Case sample identifier*|
+|`File`|`bam_or_cram_file`|Path to the GCS location of the input CRAM or BAM file.|
+
+*See **Sample ID requirements** below for specifications.
+
+The following cohort-level or batch-level inputs are also required:
+
+|Input Type|Input Name|Description|
+|---------|--------|--------------|
+|`String`|`sample_set_id`|Batch identifier|
+|`String`|`sample_set_set_id`|Cohort identifier|
+|`File`|`cohort_ped_file`|Path to the GCS location of a family structure definitions file in [PED format](/docs/gs/inputs#ped-format).|
+
+### Pipeline outputs
+
+The following are the main pipeline outputs. For more information on the outputs of each module, refer to the
+[Modules section](/docs/category/modules).
+
+|Output Type|Output Name|Description|
+|---------|--------|--------------|
+|`File`|`annotated_vcf`|Annotated SV VCF for the cohort***|
+|`File`|`annotated_vcf_idx`|Index for `annotated_vcf`|
+|`File`|`sv_vcf_qc_output`|QC plots (bundled in a .tar.gz file)|
+
+***Note that this VCF is not filtered
+
+### Pipeline overview
+
+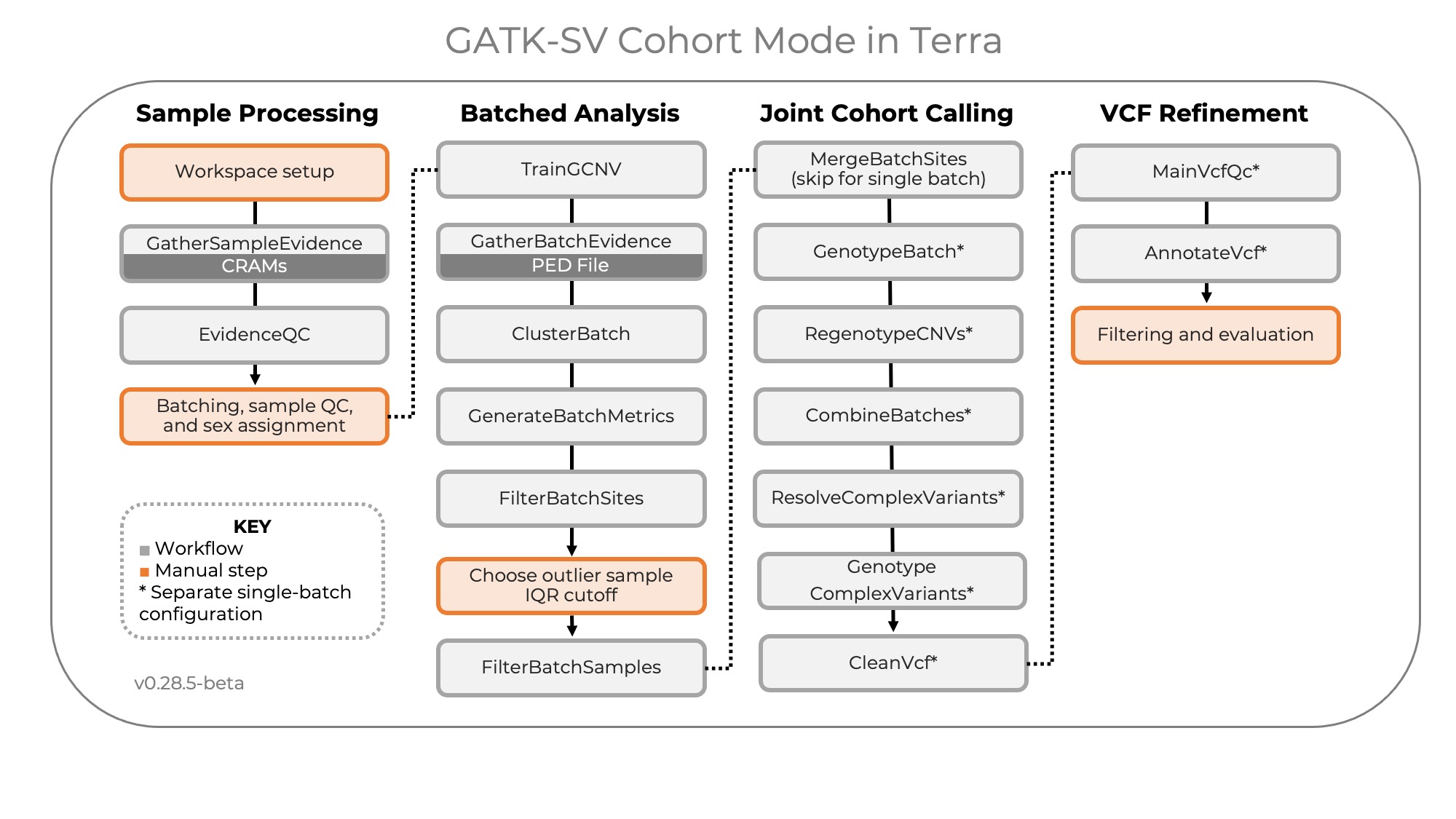 +
+The following workflows are included in this workspace, to be executed in this order:
+
+1. `01-GatherSampleEvidence`: Per-sample SV evidence collection, including calls from a configurable set of
+algorithms (Manta, MELT, and Wham), read depth (RD), split read positions (SR), and discordant pair positions (PE).
+2. `02-EvidenceQC`: Dosage bias scoring and ploidy estimation, run on preliminary batches
+3. `03-TrainGCNV`: Per-batch training of a gCNV model for use in `04-GatherBatchEvidence`
+4. `04-GatherBatchEvidence`: Per-batch copy number variant calling using cn.MOPS and GATK gCNV; B-allele frequency (BAF)
+generation; call and evidence aggregation
+5. `05-ClusterBatch`: Per-batch variant clustering
+6. `06-GenerateBatchMetrics`: Per-batch variant filtering, metric generation
+7. `07-FilterBatchSites`: Per-batch variant filtering and plot SV counts per sample per SV type to enable choice of IQR
+cutoff for outlier filtration in `08-FilterBatchSamples`
+8. `08-FilterBatchSamples`: Per-batch outlier sample filtration
+9. `09-MergeBatchSites`: Site merging of SVs discovered across batches, run on a cohort-level `sample_set_set`
+10. `10-GenotypeBatch`: Per-batch genotyping of all sites in the cohort
+11. `11-RegenotypeCNVs`: Cohort-level genotype refinement of some depth calls
+12. `12-CombineBatches`: Cohort-level cross-batch integration and clustering
+13. `13-ResolveComplexVariants`: Complex variant resolution
+14. `14-GenotypeComplexVariants`: Complex variant re-genotyping
+15. `15-CleanVcf`: VCF cleanup
+16. `16-RefineComplexVariants`: Complex variant filtering and refinement
+17. `17-ApplyManualVariantFilter`: Hard filtering high-FP SV classes
+18. `18-JoinRawCalls`: Raw call aggregation
+19. `19-SVConcordance`: Annotate genotype concordance with raw calls
+20. `20-FilterGenotypes`: Genotype filtering
+21. `21-AnnotateVcf`: Cohort VCF annotations, including functional annotation, allele frequency (AF) annotation, and
+AF annotation with external population callsets
+
+Extra workflows (Not part of canonical pipeline, but included for your convenience. May require manual configuration):
+* `MainVcfQc`: Generate detailed call set QC plots
+* `PlotSVCountsPerSample`: Plot SV counts per sample per SV type. Recommended to run before `FilterOutlierSamples`
+ (configured with the single VCF you want to filter) to enable IQR cutoff choice.
+* `FilterOutlierSamples`: Filter outlier samples (in terms of SV counts) from a single VCF.
+* `VisualizeCnvs`: Plot multi-sample depth profiles for CNVs
+
+For detailed instructions on running the pipeline in Terra, see [workflow instructions](#instructions) below.
+
+#### What is the maximum number of samples the pipeline can handle?
+
+In Terra, we have tested batch sizes of up to 500 samples and cohort sizes (consisting of multiple batches) of up to
+11,000 samples (and 98,000 samples with the final steps split by chromosome). On a dedicated Cromwell server, we have
+tested the pipeline on cohorts of up to ~140,000 samples.
+
+
+### Time and cost estimates
+
+The following estimates pertain to the 1000 Genomes sample data in this workspace. They represent aggregated run time
+and cost across modules for the whole pipeline. For workflows run multiple times (on each sample or on each batch),
+the longest individual runtime was used. Call caching may affect some of this information.
+
+|Number of samples|Time|Total run cost|Per-sample run cost|
+|--------------|--------|----------|----------|
+|312|~76 hours|~$675|~$2.16/sample|
+
+Please note that sample characteristics, cohort size, and level of filtering may influence pipeline compute costs,
+with average costs ranging between $2-$3 per sample. For instance, PCR+ samples and samples with a high percentage
+of improperly paired reads have been observed to cost more. Consider
+[excluding low-quality samples](/docs/gs/inputs#sample-exclusion) prior to processing to keep costs low.
+
+### Sample ID format
+
+Refer to [the sample ID requirements section](/docs/gs/inputs#sampleids) of the documentation.
+
+### Workspace setup
+
+1. Clone this workspace into a Terra project to which you have access
+
+2. In your new workspace, delete the example data. To do this, go to the *Data* tab of the workspace. Delete the data
+ tables in this order: `sample_set_set`, `sample_set`, and `sample`. For each table, click the 3 dots icon to the
+ right of the table name and click "Delete table". Confirm when prompted.
+
+
+The following workflows are included in this workspace, to be executed in this order:
+
+1. `01-GatherSampleEvidence`: Per-sample SV evidence collection, including calls from a configurable set of
+algorithms (Manta, MELT, and Wham), read depth (RD), split read positions (SR), and discordant pair positions (PE).
+2. `02-EvidenceQC`: Dosage bias scoring and ploidy estimation, run on preliminary batches
+3. `03-TrainGCNV`: Per-batch training of a gCNV model for use in `04-GatherBatchEvidence`
+4. `04-GatherBatchEvidence`: Per-batch copy number variant calling using cn.MOPS and GATK gCNV; B-allele frequency (BAF)
+generation; call and evidence aggregation
+5. `05-ClusterBatch`: Per-batch variant clustering
+6. `06-GenerateBatchMetrics`: Per-batch variant filtering, metric generation
+7. `07-FilterBatchSites`: Per-batch variant filtering and plot SV counts per sample per SV type to enable choice of IQR
+cutoff for outlier filtration in `08-FilterBatchSamples`
+8. `08-FilterBatchSamples`: Per-batch outlier sample filtration
+9. `09-MergeBatchSites`: Site merging of SVs discovered across batches, run on a cohort-level `sample_set_set`
+10. `10-GenotypeBatch`: Per-batch genotyping of all sites in the cohort
+11. `11-RegenotypeCNVs`: Cohort-level genotype refinement of some depth calls
+12. `12-CombineBatches`: Cohort-level cross-batch integration and clustering
+13. `13-ResolveComplexVariants`: Complex variant resolution
+14. `14-GenotypeComplexVariants`: Complex variant re-genotyping
+15. `15-CleanVcf`: VCF cleanup
+16. `16-RefineComplexVariants`: Complex variant filtering and refinement
+17. `17-ApplyManualVariantFilter`: Hard filtering high-FP SV classes
+18. `18-JoinRawCalls`: Raw call aggregation
+19. `19-SVConcordance`: Annotate genotype concordance with raw calls
+20. `20-FilterGenotypes`: Genotype filtering
+21. `21-AnnotateVcf`: Cohort VCF annotations, including functional annotation, allele frequency (AF) annotation, and
+AF annotation with external population callsets
+
+Extra workflows (Not part of canonical pipeline, but included for your convenience. May require manual configuration):
+* `MainVcfQc`: Generate detailed call set QC plots
+* `PlotSVCountsPerSample`: Plot SV counts per sample per SV type. Recommended to run before `FilterOutlierSamples`
+ (configured with the single VCF you want to filter) to enable IQR cutoff choice.
+* `FilterOutlierSamples`: Filter outlier samples (in terms of SV counts) from a single VCF.
+* `VisualizeCnvs`: Plot multi-sample depth profiles for CNVs
+
+For detailed instructions on running the pipeline in Terra, see [workflow instructions](#instructions) below.
+
+#### What is the maximum number of samples the pipeline can handle?
+
+In Terra, we have tested batch sizes of up to 500 samples and cohort sizes (consisting of multiple batches) of up to
+11,000 samples (and 98,000 samples with the final steps split by chromosome). On a dedicated Cromwell server, we have
+tested the pipeline on cohorts of up to ~140,000 samples.
+
+
+### Time and cost estimates
+
+The following estimates pertain to the 1000 Genomes sample data in this workspace. They represent aggregated run time
+and cost across modules for the whole pipeline. For workflows run multiple times (on each sample or on each batch),
+the longest individual runtime was used. Call caching may affect some of this information.
+
+|Number of samples|Time|Total run cost|Per-sample run cost|
+|--------------|--------|----------|----------|
+|312|~76 hours|~$675|~$2.16/sample|
+
+Please note that sample characteristics, cohort size, and level of filtering may influence pipeline compute costs,
+with average costs ranging between $2-$3 per sample. For instance, PCR+ samples and samples with a high percentage
+of improperly paired reads have been observed to cost more. Consider
+[excluding low-quality samples](/docs/gs/inputs#sample-exclusion) prior to processing to keep costs low.
+
+### Sample ID format
+
+Refer to [the sample ID requirements section](/docs/gs/inputs#sampleids) of the documentation.
+
+### Workspace setup
+
+1. Clone this workspace into a Terra project to which you have access
+
+2. In your new workspace, delete the example data. To do this, go to the *Data* tab of the workspace. Delete the data
+ tables in this order: `sample_set_set`, `sample_set`, and `sample`. For each table, click the 3 dots icon to the
+ right of the table name and click "Delete table". Confirm when prompted.
+  +
+3. Create and upload a new sample data table for your samples. This should be a tab-separated file (.tsv) with one line
+ per sample, as well as a header (first) line. It should contain the columns `entity:sample_id` (first column) and
+ `bam_or_cram_file` at minimum. See the **Required inputs** section above for more information on these inputs. For
+ an example sample data table, refer to the sample data table for the 1000 Genomes samples in this workspace
+ [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/samples_1kgp.tsv.tmpl).
+ To upload the TSV file, navigate to the *Data* tab of the workspace, click the `Import Data` button on the top left,
+ and select "Upload TSV".
+
+
+3. Create and upload a new sample data table for your samples. This should be a tab-separated file (.tsv) with one line
+ per sample, as well as a header (first) line. It should contain the columns `entity:sample_id` (first column) and
+ `bam_or_cram_file` at minimum. See the **Required inputs** section above for more information on these inputs. For
+ an example sample data table, refer to the sample data table for the 1000 Genomes samples in this workspace
+ [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/samples_1kgp.tsv.tmpl).
+ To upload the TSV file, navigate to the *Data* tab of the workspace, click the `Import Data` button on the top left,
+ and select "Upload TSV".
+  +
+4. Edit the `cohort_ped_file` item in the Workspace Data table (as shown in the screenshot below) to provide the Google
+ URI to the PED file for your cohort (make sure to share it with your Terra proxy account!).
+
+
+4. Edit the `cohort_ped_file` item in the Workspace Data table (as shown in the screenshot below) to provide the Google
+ URI to the PED file for your cohort (make sure to share it with your Terra proxy account!).
+  +
+
+### Creating sample_sets
+
+To create batches (in the `sample_set` table), the easiest way is to upload a tab-separated sample set membership file.
+This file should have one line per sample, plus a header (first) line. The first column should be
+`membership:sample_set_id` (containing the `sample_set_id` for the sample in question), and the second should be
+`sample` (containing the sample IDs). Recall that batch IDs (`sample_set_id`) should follow the
+[sample ID requirements](/docs/gs/inputs#sampleids). For an example sample membership file, refer to the one for the
+1000 Genomes samples in this workspace [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/sample_set_membership_1kgp.tsv.tmpl).
+
+## Workflow instructions {#instructions}
+
+### General recommendations
+
+* It is recommended to run each workflow first on one sample/batch to check that the method is properly configured
+before you attempt to process all of your data.
+* We recommend enabling call-caching (on by default in each workflow configuration).
+* We recommend enabling automatic intermediate file deletion by checking the box labeled "Delete intermediate outputs"
+at the top of the workflow launch page every time you start a workflow. With this option enabled, intermediate files
+(those not present in the Terra data table, and not needed for any further GATK-SV processing) will be deleted
+automatically if the workflow succeeds. If the workflow fails, the outputs will be retained to enable a re-run to
+pick up where it left off with call-caching. However, call-caching will not be possible for workflows that have
+succeeded. For more information on this option, see
+[this article](https://terra.bio/delete-intermediates-option-now-available-for-workflows-in-terra/). For guidance on
+managing intermediate storage from failed workflows, or from workflows without the delete intermediates option enabled,
+see the next bullet point.
+* There are cases when you may need to manage storage in other ways: for workflows that failed (only delete files from
+a failed workflow after a version has succeeded, to avoid disabling call-caching), for workflows without intermediate
+file deletion enabled, or once you are done processing and want to delete files from earlier steps in the pipeline
+that you no longer need.
+ * One option is to manually delete large files, or directories containing failed workflow intermediates (after
+ re-running the workflow successfully to take advantage of call-caching) with the command
+ `gsutil -m rm gs://path/to/workflow/directory/**file_extension_to_delete` to delete all files with the given extension
+ for that workflow, or `gsutil -m rm -r gs://path/to/workflow/directory/` to delete an entire workflow directory
+ (only after you are done with all the files!). Note that this can take a very long time for larger workflows, which
+ may contain thousands of files.
+ * Another option is to use the `fiss mop` API call to delete all files that do not appear in one of the Terra data
+ tables (intermediate files). Always ensure that you are completely done with a step and you will not need to return
+ before using this option, as it will break call-caching. See
+ [this blog post](https://terra.bio/deleting-intermediate-workflow-outputs/) for more details. This can also be done
+ [via the command line](https://github.com/broadinstitute/fiss/wiki/MOP:-reducing-your-cloud-storage-footprint).
+* If your workflow fails, check the job manager for the error message. Most issues can be resolved by increasing the
+memory or disk. Do not delete workflow log files until you are done troubleshooting. If call-caching is enabled, do not
+delete any files from the failed workflow until you have run it successfully.
+* To display run costs, see [this article](https://support.terra.bio/hc/en-us/articles/360037862771#h_01EX5ED53HAZ59M29DRCG24CXY)
+for one-time setup instructions for non-Broad users.
+
+### 01-GatherSampleEvidence
+
+Read the full GatherSampleEvidence documentation [here](/docs/modules/gse).
+* This workflow runs on a per-sample level, but you can launch many (a few hundred) samples at once, in arbitrary
+partitions. Make sure to try just one sample first though!
+* Refer to the [Input Data section](/docs/gs/inputs) for details on file formats, sample QC, and sample ID restrictions.
+* It is normal for a few samples in a cohort to run out of memory during Wham SV calling, so we recommend enabling
+auto-retry for out-of-memory errors for `01-GatherSampleEvidence` only. Before you launch the workflow, click the
+checkbox reading "Retry with more memory" and set the memory retry factor to 1.8. This action must be performed each
+time you launch a `01-GatherSampleEvidence` job.
+* Please note that most large published joint call sets produced by GATK-SV, including gnomAD-SV, included the tool
+MELT, a state-of-the-art mobile element insertion (MEI) detector, as part of the pipeline. Due to licensing
+restrictions, we cannot provide a public docker image for this algorithm. The `01-GatherSampleEvidence` workflow
+does not use MELT as one of the SV callers by default, which will result in less sensitivity to MEI calls. In order
+to use MELT, you will need to build your own private docker image (example Dockerfile
+[here](https://github.com/broadinstitute/gatk-sv/blob/master/dockerfiles/melt/Dockerfile)), share it with your Terra
+proxy account, enter it in the `melt_docker` input in the `01-GatherSampleEvidence` configuration (as a string,
+surrounded by double-quotes), and then click "Save". No further changes are necessary beyond `01-GatherSampleEvidence`.
+ * Note that the version of MELT tested with GATK-SV is v2.0.5. If you use a different version to create your own
+ docker image, we recommend testing your image by running one pilot sample through `01-GatherSampleEvidence` to check
+ that it runs as expected, then running a small group of about 10 pilot samples through the pipeline until the end of
+ `04-GatherBatchEvidence` to check that the outputs are compatible with GATK-SV.
+* If you enable "Delete intermediate outputs" whenever you launch this workflow (recommended), BAM files will be
+deleted for successful runs; but BAM files will not be deleted if the run fails or if intermediate file deletion is
+not enabled. Since BAM files are large, we recommend deleting them to save on storage costs, but only after fixing and
+re-running the failed workflow, so that it will call-cache.
+
+
+### 02-EvidenceQC
+
+Read the full EvidenceQC documentation [here](/docs/modules/eqc).
+* `02-EvidenceQC` is run on arbitrary cohort partitions of up to 500 samples.
+* The outputs from `02-EvidenceQC` can be used for
+[preliminary sample QC](/docs/modules/eqc#preliminary-sample-qc) and
+[batching](#batching) before moving on to [TrainGCNV](#traingcnv).
+
+
+### Batching (manual step) {#batching}
+
+For larger cohorts, samples should be split up into batches of about 100-500
+samples with similar characteristics. We recommend batching based on overall
+coverage and dosage score (WGD), which can be generated in [EvidenceQC](/docs/modules/eqc).
+An example batching process is outlined below:
+
+1. Divide the cohort into PCR+ and PCR- samples
+2. Partition the samples by median coverage from [EvidenceQC](/docs/modules/eqc),
+ grouping samples with similar median coverage together. The end goal is to
+ divide the cohort into roughly equal-sized batches of about 100-500 samples;
+ if your partitions based on coverage are larger or uneven, you can partition
+ the cohort further in the next step to obtain the final batches.
+3. Optionally, divide the samples further by dosage score (WGD) from
+ [EvidenceQC](/docs/modules/eqc), grouping samples with similar WGD score
+ together, to obtain roughly equal-sized batches of about 100-500 samples
+4. Maintain a roughly equal sex balance within each batch, based on sex
+ assignments from [EvidenceQC](/docs/modules/eqc)
+
+
+### 03-TrainGCNV {#traingcnv}
+
+Read the full TrainGCNV documentation [here](/docs/modules/gcnv).
+* Before running this workflow, create the batches (~100-500 samples) you will use for the rest of the pipeline based
+on sample coverage, WGD score (from `02-EvidenceQC`), and PCR status. These will likely not be the same as the batches
+you used for `02-EvidenceQC`.
+* By default, `03-TrainGCNV` is configured to be run once per `sample_set` on 100 randomly-chosen samples from that
+set to create a gCNV model for each batch. To modify this behavior, you can set the `n_samples_subsample` parameter
+to the number of samples to use for training.
+
+### 04-GatherBatchEvidence
+
+Read the full GatherBatchEvidence documentation [here](/docs/modules/gbe).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV`.
+* Before running this workflow, ensure that you have updated the `cohort_ped_file` attribute in Workspace Data with
+your cohort's PED file, with sex assignments updated based on ploidy detection from `02-EvidenceQC`.
+
+### Steps 05-06
+
+Read the full documentation for [ClusterBatch](/docs/modules/cb) and [GenerateBatchMetrics](/docs/modules/gbm).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` and `04-GatherBatchEvidence`.
+
+
+### Steps 07-08
+
+These two workflows make up FilterBatch; they are subdivided in this workspace to enable tuning of outlier filtration
+cutoffs. Read the full FilterBatch documentation [here](/docs/modules/fb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `06-GenerateBatchMetrics`.
+* `07-FilterBatchSites` produces SV count plots and files, as well as a preview of the outlier samples to be filtered.
+The input `N_IQR_cutoff_plotting` is used to visualize filtration thresholds on the SV count plots and preview the
+samples to be filtered; the default value is set to 6. You can adjust this value depending on your needs, and you can
+re-run the workflow with new `N_IQR_cutoff_plotting` values until the plots and outlier sample lists suit the purposes
+of your study. Once you have chosen an IQR cutoff, provide it to the `N_IQR_cutoff` input in `08-FilterBatchSamples` to
+filter the VCFs using the chosen cutoff.
+* `08-FilterBatchSamples` performs outlier sample filtration, removing samples with an abnormal number of SV calls of
+at least one SV type. To tune the filtering threshold to your needs, edit the `N_IQR_cutoff` input value based on the
+plots and outlier sample preview lists from `07-FilterBatchSites`. The default value for `N_IQR_cutoff` in this step
+is 10000, which essentially means that no samples are filtered.
+
+### 09-MergeBatchSites
+
+Read the full MergeBatchSites documentation [here](/docs/modules/msites).
+* `09-MergeBatchSites` is a cohort-level workflow, so it is run on a `sample_set_set` containing all the batches
+in the cohort. Navigate to the Data tab of your workspace. If there is no `sample_set_set` data table, you will need
+to create it. To do this, select the `sample_set` data table, then select (with the checkboxes) all the batches
+(`sample_set`) in your cohort. These should be the `sample_sets` that you used to run steps `03-TrainGCNV` through
+`08-FilterBatchSamples`. Then click the "Edit" icon above the table and choose "Save selection as set." Enter a name
+that follows the **Sample ID requirements**. This will create a new `sample_set_set` containing all of the `sample_sets`
+in your cohort. When you launch MergeBatchSites, you can now select this `sample_set_set`.
+
+
+
+
+### Creating sample_sets
+
+To create batches (in the `sample_set` table), the easiest way is to upload a tab-separated sample set membership file.
+This file should have one line per sample, plus a header (first) line. The first column should be
+`membership:sample_set_id` (containing the `sample_set_id` for the sample in question), and the second should be
+`sample` (containing the sample IDs). Recall that batch IDs (`sample_set_id`) should follow the
+[sample ID requirements](/docs/gs/inputs#sampleids). For an example sample membership file, refer to the one for the
+1000 Genomes samples in this workspace [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/sample_set_membership_1kgp.tsv.tmpl).
+
+## Workflow instructions {#instructions}
+
+### General recommendations
+
+* It is recommended to run each workflow first on one sample/batch to check that the method is properly configured
+before you attempt to process all of your data.
+* We recommend enabling call-caching (on by default in each workflow configuration).
+* We recommend enabling automatic intermediate file deletion by checking the box labeled "Delete intermediate outputs"
+at the top of the workflow launch page every time you start a workflow. With this option enabled, intermediate files
+(those not present in the Terra data table, and not needed for any further GATK-SV processing) will be deleted
+automatically if the workflow succeeds. If the workflow fails, the outputs will be retained to enable a re-run to
+pick up where it left off with call-caching. However, call-caching will not be possible for workflows that have
+succeeded. For more information on this option, see
+[this article](https://terra.bio/delete-intermediates-option-now-available-for-workflows-in-terra/). For guidance on
+managing intermediate storage from failed workflows, or from workflows without the delete intermediates option enabled,
+see the next bullet point.
+* There are cases when you may need to manage storage in other ways: for workflows that failed (only delete files from
+a failed workflow after a version has succeeded, to avoid disabling call-caching), for workflows without intermediate
+file deletion enabled, or once you are done processing and want to delete files from earlier steps in the pipeline
+that you no longer need.
+ * One option is to manually delete large files, or directories containing failed workflow intermediates (after
+ re-running the workflow successfully to take advantage of call-caching) with the command
+ `gsutil -m rm gs://path/to/workflow/directory/**file_extension_to_delete` to delete all files with the given extension
+ for that workflow, or `gsutil -m rm -r gs://path/to/workflow/directory/` to delete an entire workflow directory
+ (only after you are done with all the files!). Note that this can take a very long time for larger workflows, which
+ may contain thousands of files.
+ * Another option is to use the `fiss mop` API call to delete all files that do not appear in one of the Terra data
+ tables (intermediate files). Always ensure that you are completely done with a step and you will not need to return
+ before using this option, as it will break call-caching. See
+ [this blog post](https://terra.bio/deleting-intermediate-workflow-outputs/) for more details. This can also be done
+ [via the command line](https://github.com/broadinstitute/fiss/wiki/MOP:-reducing-your-cloud-storage-footprint).
+* If your workflow fails, check the job manager for the error message. Most issues can be resolved by increasing the
+memory or disk. Do not delete workflow log files until you are done troubleshooting. If call-caching is enabled, do not
+delete any files from the failed workflow until you have run it successfully.
+* To display run costs, see [this article](https://support.terra.bio/hc/en-us/articles/360037862771#h_01EX5ED53HAZ59M29DRCG24CXY)
+for one-time setup instructions for non-Broad users.
+
+### 01-GatherSampleEvidence
+
+Read the full GatherSampleEvidence documentation [here](/docs/modules/gse).
+* This workflow runs on a per-sample level, but you can launch many (a few hundred) samples at once, in arbitrary
+partitions. Make sure to try just one sample first though!
+* Refer to the [Input Data section](/docs/gs/inputs) for details on file formats, sample QC, and sample ID restrictions.
+* It is normal for a few samples in a cohort to run out of memory during Wham SV calling, so we recommend enabling
+auto-retry for out-of-memory errors for `01-GatherSampleEvidence` only. Before you launch the workflow, click the
+checkbox reading "Retry with more memory" and set the memory retry factor to 1.8. This action must be performed each
+time you launch a `01-GatherSampleEvidence` job.
+* Please note that most large published joint call sets produced by GATK-SV, including gnomAD-SV, included the tool
+MELT, a state-of-the-art mobile element insertion (MEI) detector, as part of the pipeline. Due to licensing
+restrictions, we cannot provide a public docker image for this algorithm. The `01-GatherSampleEvidence` workflow
+does not use MELT as one of the SV callers by default, which will result in less sensitivity to MEI calls. In order
+to use MELT, you will need to build your own private docker image (example Dockerfile
+[here](https://github.com/broadinstitute/gatk-sv/blob/master/dockerfiles/melt/Dockerfile)), share it with your Terra
+proxy account, enter it in the `melt_docker` input in the `01-GatherSampleEvidence` configuration (as a string,
+surrounded by double-quotes), and then click "Save". No further changes are necessary beyond `01-GatherSampleEvidence`.
+ * Note that the version of MELT tested with GATK-SV is v2.0.5. If you use a different version to create your own
+ docker image, we recommend testing your image by running one pilot sample through `01-GatherSampleEvidence` to check
+ that it runs as expected, then running a small group of about 10 pilot samples through the pipeline until the end of
+ `04-GatherBatchEvidence` to check that the outputs are compatible with GATK-SV.
+* If you enable "Delete intermediate outputs" whenever you launch this workflow (recommended), BAM files will be
+deleted for successful runs; but BAM files will not be deleted if the run fails or if intermediate file deletion is
+not enabled. Since BAM files are large, we recommend deleting them to save on storage costs, but only after fixing and
+re-running the failed workflow, so that it will call-cache.
+
+
+### 02-EvidenceQC
+
+Read the full EvidenceQC documentation [here](/docs/modules/eqc).
+* `02-EvidenceQC` is run on arbitrary cohort partitions of up to 500 samples.
+* The outputs from `02-EvidenceQC` can be used for
+[preliminary sample QC](/docs/modules/eqc#preliminary-sample-qc) and
+[batching](#batching) before moving on to [TrainGCNV](#traingcnv).
+
+
+### Batching (manual step) {#batching}
+
+For larger cohorts, samples should be split up into batches of about 100-500
+samples with similar characteristics. We recommend batching based on overall
+coverage and dosage score (WGD), which can be generated in [EvidenceQC](/docs/modules/eqc).
+An example batching process is outlined below:
+
+1. Divide the cohort into PCR+ and PCR- samples
+2. Partition the samples by median coverage from [EvidenceQC](/docs/modules/eqc),
+ grouping samples with similar median coverage together. The end goal is to
+ divide the cohort into roughly equal-sized batches of about 100-500 samples;
+ if your partitions based on coverage are larger or uneven, you can partition
+ the cohort further in the next step to obtain the final batches.
+3. Optionally, divide the samples further by dosage score (WGD) from
+ [EvidenceQC](/docs/modules/eqc), grouping samples with similar WGD score
+ together, to obtain roughly equal-sized batches of about 100-500 samples
+4. Maintain a roughly equal sex balance within each batch, based on sex
+ assignments from [EvidenceQC](/docs/modules/eqc)
+
+
+### 03-TrainGCNV {#traingcnv}
+
+Read the full TrainGCNV documentation [here](/docs/modules/gcnv).
+* Before running this workflow, create the batches (~100-500 samples) you will use for the rest of the pipeline based
+on sample coverage, WGD score (from `02-EvidenceQC`), and PCR status. These will likely not be the same as the batches
+you used for `02-EvidenceQC`.
+* By default, `03-TrainGCNV` is configured to be run once per `sample_set` on 100 randomly-chosen samples from that
+set to create a gCNV model for each batch. To modify this behavior, you can set the `n_samples_subsample` parameter
+to the number of samples to use for training.
+
+### 04-GatherBatchEvidence
+
+Read the full GatherBatchEvidence documentation [here](/docs/modules/gbe).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV`.
+* Before running this workflow, ensure that you have updated the `cohort_ped_file` attribute in Workspace Data with
+your cohort's PED file, with sex assignments updated based on ploidy detection from `02-EvidenceQC`.
+
+### Steps 05-06
+
+Read the full documentation for [ClusterBatch](/docs/modules/cb) and [GenerateBatchMetrics](/docs/modules/gbm).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` and `04-GatherBatchEvidence`.
+
+
+### Steps 07-08
+
+These two workflows make up FilterBatch; they are subdivided in this workspace to enable tuning of outlier filtration
+cutoffs. Read the full FilterBatch documentation [here](/docs/modules/fb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `06-GenerateBatchMetrics`.
+* `07-FilterBatchSites` produces SV count plots and files, as well as a preview of the outlier samples to be filtered.
+The input `N_IQR_cutoff_plotting` is used to visualize filtration thresholds on the SV count plots and preview the
+samples to be filtered; the default value is set to 6. You can adjust this value depending on your needs, and you can
+re-run the workflow with new `N_IQR_cutoff_plotting` values until the plots and outlier sample lists suit the purposes
+of your study. Once you have chosen an IQR cutoff, provide it to the `N_IQR_cutoff` input in `08-FilterBatchSamples` to
+filter the VCFs using the chosen cutoff.
+* `08-FilterBatchSamples` performs outlier sample filtration, removing samples with an abnormal number of SV calls of
+at least one SV type. To tune the filtering threshold to your needs, edit the `N_IQR_cutoff` input value based on the
+plots and outlier sample preview lists from `07-FilterBatchSites`. The default value for `N_IQR_cutoff` in this step
+is 10000, which essentially means that no samples are filtered.
+
+### 09-MergeBatchSites
+
+Read the full MergeBatchSites documentation [here](/docs/modules/msites).
+* `09-MergeBatchSites` is a cohort-level workflow, so it is run on a `sample_set_set` containing all the batches
+in the cohort. Navigate to the Data tab of your workspace. If there is no `sample_set_set` data table, you will need
+to create it. To do this, select the `sample_set` data table, then select (with the checkboxes) all the batches
+(`sample_set`) in your cohort. These should be the `sample_sets` that you used to run steps `03-TrainGCNV` through
+`08-FilterBatchSamples`. Then click the "Edit" icon above the table and choose "Save selection as set." Enter a name
+that follows the **Sample ID requirements**. This will create a new `sample_set_set` containing all of the `sample_sets`
+in your cohort. When you launch MergeBatchSites, you can now select this `sample_set_set`.
+
+ +
+ +* If there is already a `sample_set_set` data table in your workspace, you can create this `sample_set_set` while you
+are launching the `09-MergeBatchSites` workflow: click "Select Data", choose "Create new sample_set_set [...]", check
+all the batches to include (all the ones used in `03-TrainGCNV` through `08-FilterBatchSamples`), and give it a name
+that follows the [sample ID requirements](/docs/gs/inputs#sampleids).
+
+
+* If there is already a `sample_set_set` data table in your workspace, you can create this `sample_set_set` while you
+are launching the `09-MergeBatchSites` workflow: click "Select Data", choose "Create new sample_set_set [...]", check
+all the batches to include (all the ones used in `03-TrainGCNV` through `08-FilterBatchSamples`), and give it a name
+that follows the [sample ID requirements](/docs/gs/inputs#sampleids).
+
+ +
+### 10-GenotypeBatch
+
+Read the full GenotypeBatch documentation [here](/docs/modules/gb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `08-FilterBatchSamples`.
+
+### Steps 11-17
+
+Read the full documentation for [RegenotypeCNVs](/docs/modules/rgcnvs), [MakeCohortVcf](/docs/modules/cvcf) (which
+includes `CombineBatches`, `ResolveComplexVariants`, `GenotypeComplexVariants`, `CleanVcf`, `MainVcfQc`), and
+[AnnotateVcf](/docs/modules/av).
+* Use the same cohort `sample_set_set` you created and used for `09-MergeBatchSites`.
+
+### Additional notes
+
+- The VCF produced by `15-CleanVcf` (and annotated by `17-AnnotateVcf`) prioritizes sensitivity, but additional downstream
+filtration is recommended to improve specificity.
+
diff --git a/website/docs/execution/overview.md b/website/docs/execution/overview.md
new file mode 100644
index 000000000..fecfd3825
--- /dev/null
+++ b/website/docs/execution/overview.md
@@ -0,0 +1,10 @@
+---
+title: Overview
+description: Overview
+sidebar_position: 1
+slug: overview
+---
+
+This section provides technical documentation for the running single-sample and joint calling modes in Terra.
+
+Please review the [Getting Started](/docs/gs/overview) material before proceeding with this section.
diff --git a/website/docs/execution/single.md b/website/docs/execution/single.md
new file mode 100644
index 000000000..1132a2ac3
--- /dev/null
+++ b/website/docs/execution/single.md
@@ -0,0 +1,121 @@
+---
+title: Single-sample
+description: Run the pipeline on a single sample
+sidebar_position: 3
+slug: single
+---
+
+## Introduction
+
+**Extending SV detection to small datasets**
+
+The Single Sample pipeline is designed to facilitate running the methods developed for the cohort-mode GATK-SV pipeline on small data sets or in
+clinical contexts where batching large numbers of samples is not an option. To do so, it uses precomputed data, SV calls,
+and model parameters computed by the cohort pipeline on a reference panel composed of similar samples. The pipeline integrates this
+precomputed information with signals extracted from the input CRAM file to produce a call set similar in quality to results
+computed by the cohort pipeline in a computationally tractable and reproducible manner.
+
+
+## Terra workspace
+Users should clone the [Terra single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+which is configured with a demo sample to run on.
+Refer to the following sections for instructions on how to run the pipeline on your data using this workspace.
+
+## Data
+
+### Case sample
+
+The default workspace includes a NA12878 input WGS CRAM file configured in the workspace samples data table. This file is part of the
+high coverage (30X) [WGS data](https://www.internationalgenome.org/data-portal/data-collection/30x-grch38) for the 1000
+Genomes Project samples generated by the New York Genome Center and hosted in
+[AnVIL](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019).
+
+### Reference panel
+
+The reference panel configured in the workspace consists of data and calls computed from 156 publicly available samples
+chosen from the NYGC/AnVIL 1000 Genomes high coverage data linked above.
+
+Inputs to the pipeline for the reference panel include:
+- A precomputed SV callset VCF, and joint-called depth-based CNV call files
+- Raw calls for the reference panel samples from Manta and WHAM
+- Trained models for calling copy number variation in GATK gCNV case mode
+- Parameters learned by the cohort mode pipeline in training machine learning models on the reference panel samples.
+
+These resources are primarily configured in the "Workspace Data" for this workspace. However, several of the resources need
+to be passed to the workflow as large lists of files or strings. Due to Terra limitations on uploading data containing lists to the
+workspace data table, these resources are specified directly in the workflow configuration.
+
+### Reference resources
+
+The pipeline uses a number of resource and data files computed for the hg38 reference:
+- Reference sequences and indices
+- Genome annotation tracks such as segmental duplication and RepeatMasker tracks
+- Data used for annotation of called variants, including GenCode gene annotations and gnomAD site allele frequencies.
+
+## Single sample workflow
+
+### What does it do?
+
+The workflow `gatk-sv-single-sample` calls structural variations on a single input CRAM by running the GATK-SV Single Sample Pipeline end-to-end.
+
+#### What does it require as input?
+
+The workflow accepts a single CRAM or BAM file as input, configured in the following parameters:
+
+|Input Type|Input Name|Description|
+|---------|--------|--------------|
+|`String`|`sample_id`|Case sample identifier|
+|`File`|`bam_or_cram_file`|Path to the GCS location of the input CRAM or BAM file|
+|`String`|`batch`|Arbitrary name to be assigned to the run|
+
+#### Additional workspace-level inputs
+
+- Reference resources for hg38
+- Input data for the reference panel
+- The set of docker images used in the pipeline.
+
+Please contact GATK-SV developers if you are interested in customizing these
+inputs beyond their defaults.
+
+#### What does it return as output?
+
+|Output Type|Output Name|Description|
+|---------|--------|--------------|
+|`File`|`final_vcf`|SV VCF output for the pipeline. Includes all sites genotyped as variant in the case sample and genotypes for the reference panel. Sites are annotated with overlap of functional genome elements and allele frequencies of matching variants in gnomAD|
+|`File`|`final_vcf_idx`|Index file for `final_vcf`|
+|`File`|`final_bed`|Final output in BED format. Filter status, list of variant samples, and all VCF INFO fields are reported as additional columns.|
+|`File`|`metrics_file`|Metrics computed from the input data and intermediate and final VCFs. Includes metrics on the SV evidence, and on the number of variants called, broken down by type and size range.|
+|`File`|`qc_file`|Quality-control check file. This extracts several key metrics from the `metrics_file` and compares them to pre-specified threshold values. If any QC checks evaluate to FAIL, further diagnostics may be required.|
+|`File`|`ploidy_matrix`|Matrix of contig ploidy estimates computed by GATK gCNV.|
+|`File`|`ploidy_plots`|Plots of contig ploidy generated from `ploidy_matrix`|
+|`File`|`non_genotyped_unique_depth_calls`|This VCF file contains any depth based calls made in the case sample that did not pass genotyping checks and do not match a depth-based call from the reference panel. If very high sensitivity is desired, examine this file for additional large CNV calls.|
+|`File`|`non_genotyped_unique_depth_calls_idx`|Index file for `non_genotyped_unique_depth_calls`|
+|`File`|`pre_cleanup_vcf`|VCF output in a representation used internally in the pipeline. This file is less compliant with the VCF spec and is intended for debugging purposes.|
+|`File`|`pre_cleanup_vcf_idx`|Index file for `pre_cleanup_vcf`|
+
+#### Example time and cost run on sample data
+
+|Sample Name|Sample Size|Time|Cost $|
+|-----------|-----------|----|------|
+|NA12878|18.17 GiB|23hrs|~$7.34|
+
+#### To use this workflow on your own data
+
+If you would like to run this workflow on your own samples (which must be medium-to-high coverage WGS data):
+
+- Clone the [workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample) into a Terra project you have access to
+- In the cloned workspace, upload rows to the Sample and (optionally) the Participant Data Table that describe your samples.
+ Ensure that the rows you add to the Sample table contain the columns `sample_id` and `bam_or_cram_file` are populated appropriately.
+- There is no need to modify values in the workspace data or method configuration. If you are interested in modifying the reference
+ genome resources or reference panel, please contact the GATK team for support as listed below.
+- Launch the workflow from the "Workflows" tab, selecting your samples as the inputs.
+
+#### Quality control
+
+Please check the `qc_file` output to screen for data quality issues. This file provides a table of quality metrics and
+suggested acceptable ranges.
+
+:::warning
+If a metric exceeds the recommended range, all variants will be automatically flagged with
+a non-passing `FILTER` status in the output VCF.
+:::
\ No newline at end of file
diff --git a/website/docs/gs/_category_.json b/website/docs/gs/_category_.json
index 6b9e1272a..fe28ad97e 100644
--- a/website/docs/gs/_category_.json
+++ b/website/docs/gs/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Getting Started",
- "position": 2,
+ "position": 3,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/gs/calling_modes.md b/website/docs/gs/calling_modes.md
new file mode 100644
index 000000000..dcafa5634
--- /dev/null
+++ b/website/docs/gs/calling_modes.md
@@ -0,0 +1,41 @@
+---
+title: Calling modes
+description: Description of single-sample and joint calling
+sidebar_position: 1
+---
+
+# Calling modes
+
+GATK-SV offers two different modes for SV calling. Users should carefully review the following sections to determine
+which mode is appropriate for their use case.
+
+## Single-sample mode
+
+GATK-SV can perform SV calling on individual samples. In this mode, a sample is jointly called against a fixed reference
+panel of [156 high-quality samples from the 1000 Genomes Project](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019). Single-sample mode is a good option for the following
+use cases:
+
+- Studies involving fewer 100 samples
+- Studies with rolling data delivery, i.e. in small batches over time
+
+Users should also consider that the single-sample mode is provided as a single workflow and is therefore considerably
+simpler to run than joint calling. However, it also has higher compute costs on a per-sample basis and will not be as sensitive
+as joint calling with larger cohorts.
+
+## Joint calling mode
+
+GATK-SV can also perform joint calling on a set of samples. Users may opt for this mode in the following use cases:
+
+- Studies involving at least 100 samples
+- When maximum sensitivity is desired
+- Data sets that are technically heterogeneous, i.e. with strong batch effects, or are very different from the single-sample mode reference panel
+
+Joint calling has the advantage of increasing SV discovery sensitivity and providing allele frequency estimates, and there are
+some features, such as genotype recalibration and filtering and in-depth QC plotting, that are only available in joint calling mode.
+However, this pipeline is considerably more complex to execute than the single-sample mode, requiring sample batching and the execution of
+several individual modules.
+
+## Related content
+
+More information on single-sample and joint calling can be found in the [Execution](/docs/execution/overview) section.
+
diff --git a/website/docs/gs/dockers.md b/website/docs/gs/dockers.md
new file mode 100644
index 000000000..7a4e0f658
--- /dev/null
+++ b/website/docs/gs/dockers.md
@@ -0,0 +1,73 @@
+---
+title: Docker images
+description: Docker images
+sidebar_position: 4
+---
+
+GATK-SV utilizes a set of [Docker](https://www.docker.com/) images for execution within containerized environments.
+
+### Publishing and availability
+
+Dockers are automatically built and pushed to the `us.gcr.io/broad-dsde-methods/gatk-sv` repository under two different conditions:
+1. **Release**: upon releasing a new version of GATK-SV. These Dockers are made permanently available.
+2. **Commit**: upon merging a new commit to the development branch. These Dockers are ephemeral and may be periodically
+deleted. Any users needing to preserve access to these Docker images should copy them to their own repository. Also
+note that these images are built on an "as-needed" basis, meaning an image is only updated if it has been changed
+in any way by the commit.
+
+The full set of current images are automatically published and pushed to a public
+[Google Artifact Registry](https://cloud.google.com/artifact-registry/docs) and listed in the
+[dockers.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers.json)
+file.
+
+:::info
+Microsoft Azure mirrors of all images can be found in
+[dockers_azure.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers_azure.json),
+but these are not currently available for public use.
+:::
+
+### Regions (IMPORTANT)
+
+Users using Google Compute VMs outside of the `us-central1` region, i.e. in other NA regions or continents, must copy all
+docker images to a repository hosted in their region. If using a Terra workspace, the region is listed under `Cloud
+Information`:`Location` in the workspace dashboard. Please see
+[this article](https://support.terra.bio/hc/en-us/articles/4408985788187-How-to-configure-Google-Artifact-Registry-to-prevent-data-transfer-egress-charges)
+for more information.
+
+:::warning
+Failure to localize Docker images to your region will incur significant egress costs.
+:::
+
+### Versioning
+
+All Docker images are tagged with a date and version number that must be run with the corresponding version of the
+WDLs. The Docker images built with a particular version can be determined from the `dockers.json` file by checking out
+the commit or release of interest and examining `dockers.json`, e.g.
+[v0.29-beta](https://github.com/broadinstitute/gatk-sv/blob/v0.29-beta/inputs/values/dockers.json).
+
+Note that a given commit may contain a mixture of Docker image version tags if only a subset of images has actually
+been updated, reflecting the "as-needed" rebuild policy described above.
+
+All Terra workspace releases have the WDLs and Docker images synchronized.
+
+:::info
+Cloning a Terra workspace copies a snapshot of the current workspace. Any future updates to the original workspace
+will not be propagated to the clone.
+:::
+
+:::warning
+We strongly recommend using a single version of GATK-SV for all projects. Switching versions may lead to batch effects
+and/or compromise call set fidelity.
+:::
+
+
+### Workflow inputs
+
+All GATK-SV WDLs utilize a subset of the Docker images, which are specified as inputs. For example,
+the `sv_pipeline` docker is supplied to the input `sv_pipeline_docker` in many WDLs.
+
+
+### Builds
+
+An in-depth guide to the GATK-SV Docker system and building Docker images is available in the
+[Advanced section](/docs/category/docker-builds).
diff --git a/website/docs/gs/input_files.md b/website/docs/gs/input_files.md
index 38e143869..aafd031f1 100644
--- a/website/docs/gs/input_files.md
+++ b/website/docs/gs/input_files.md
@@ -1,62 +1,72 @@
---
-title: Input Data
+title: Input data
description: Supported input and reference data.
-sidebar_position: 5
+sidebar_position: 2
slug: ./inputs
---
GATK-SV requires the following input data:
-- Illumina short-read whole-genome CRAMs or BAMs, aligned to hg38 with [bwa-mem](https://github.com/lh3/bwa).
- BAMs must also be indexed.
+1. Sequencing alignments in BAM or CRAM format that are:
+ - Short-read, paired-end Illumina (e.g. Novaseq)
+ - Deep whole-genome coverage (~30x); RNA-seq and targeted (exome) libraries are not supported
+ - Indexed (have a companion `.bai` or `.crai` file)
+ - Aligned to
+ [hg38](https://gatk.broadinstitute.org/hc/en-us/articles/360035890951-Human-genome-reference-builds-GRCh38-or-hg38-b37-hg19)
+ with either [GATK Best Practices](https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows)
+ and [bwa-mem](https://github.com/lh3/bwa),
+ or [Illumina DRAGEN](https://www.illumina.com/products/by-type/informatics-products/dragen-secondary-analysis.html) v3.4.12 or v3.7.8
+2. (Joint calling mode only) Family structure definitions file in [PED format](/docs/gs/inputs#ped-format). This file is required even if your dataset does not contain related individuals.
-- Family structure definitions file in
- [PED format](/docs/gs/inputs#ped-format).
-
-### PED file format {#ped-format}
-The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
-* The file must be tab-delimited.
-* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
-* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
-* Missing parental IDs should be entered as 0.
-* Header lines are allowed if they begin with a # character.
-To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
+Note that the supported alignment pipeline versions have been extensively tested for robustness and accuracy. While other
+versions of DRAGEN may work as well, they have not been validated with GATK-SV. We do not recommend mixing aligners within call sets.
### Sample Exclusion
We recommend filtering out samples with a high percentage
-of improperly paired reads (>10% or an outlier for your data)
+of improperly paired or chimeric reads
as technical outliers prior to running [GatherSampleEvidence](/docs/modules/gse).
-A high percentage of improperly paired reads may indicate issues
-with library prep, degradation, or contamination. Artifactual
-improperly paired reads could cause incorrect SV calls, and
-these samples have been observed to have longer runtimes and
-higher compute costs for [GatherSampleEvidence](/docs/modules/gse).
+Samples with high rates of anomalous reads may indicate issues
+with library preparation, degradation, or contamination and can lead to poor variant set quality.
+Samples failing these criteria often require longer run times and higher compute costs.
-### Sample ID requirements {#sampleids}
-#### Sample IDs must
+### Sample IDs {#sampleids}
+GATK-SV imposes certain restrictions on sample names (IDs) in order to avoid certain parsing errors (e.g. with the
+use of the `grep` command). While future releases will obviate some of these restrictions, users must modify
+their sample IDs according to the following requirements.
+
+#### Sample IDs must:
- Be unique within the cohort
- Contain only alphanumeric characters and underscores (no dashes, whitespace, or special characters)
-#### Sample IDs should not
-
-- Contain only numeric characters
+#### Sample IDs should not:
+- Contain only numeric characters, e.g. `10004928`
- Be a substring of another sample ID in the same cohort
- Contain any of the following substrings: `chr`, `name`, `DEL`, `DUP`, `CPX`, `CHROM`
-The same requirements apply to family IDs in the PED file,
-as well as batch IDs and the cohort ID provided as workflow inputs.
+The same requirements apply to family IDs in the PED file, as well as batch IDs and the cohort ID provided as workflow inputs.
-Sample IDs are provided to GatherSampleEvidence directly and
-need not match sample names from the BAM/CRAM headers.
-`GetSampleID.wdl` can be used to fetch BAM sample IDs and
-also generates a set of alternate IDs that are considered
-safe for this pipeline; alternatively, [this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
+Users should set sample IDs in [GatherSampleEvidence](/docs/modules/gse) with the `sample_id` input, which needs not match
+the sample name defined in the BAM/CRAM header. `GetSampleID.wdl` can be used to fetch BAM sample IDs and also generates a set
+of alternate IDs that are considered safe for this pipeline. Alternatively,
+[this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
transforms a list of sample IDs to fit these requirements.
-Currently, sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe).
-The following inputs will need to be updated with the transformed sample IDs:
+Sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe). To do so, set the parameter
+`rename_samples = True` and provide updated sample IDs via the `samples` parameter.
+
+Note that following inputs will need to be updated with the transformed sample IDs:
- Sample ID list for [GatherSampleEvidence](/docs/modules/gse) or [GatherBatchEvidence](/docs/modules/gbe)
- PED file
+
+
+### PED file format {#ped-format}
+The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
+* The file must be tab-delimited.
+* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
+* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
+* Missing parental IDs should be entered as 0.
+* Header lines are allowed if they begin with a # character.
+* To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
diff --git a/website/docs/gs/overview.md b/website/docs/gs/overview.md
index 5b147c973..f4493759c 100644
--- a/website/docs/gs/overview.md
+++ b/website/docs/gs/overview.md
@@ -6,9 +6,11 @@ sidebar_position: 0
# Overview
-GATK-SV is a highly-scalable cloud-native pipeline for structural variation discovery
-on Illumina short-read whole-genome sequencing (WGS) data.
-The pipeline genotypes structural variations using Docker-based tools, modularized in
-different components, and orchestrated using Cromwell.
+This section provides guidance on how GATK-SV can be used, restrictions on input data, and where to get started
+with generating call sets.
-Please refer to the documentation on [this page](/docs/run/overview) for running the pipeline.
+We highly recommend that users new to SV calling also review the [Best Practices Guide](/docs/best_practices).
+
+Most users should use the Terra platform for running GATK-SV and, after reading this section,
+should refer to the documentation in the [Execution section](/docs/execution/overview) for instructions on
+how to run their own data.
diff --git a/website/docs/gs/runtime_env.md b/website/docs/gs/runtime_env.md
index 65aee6032..cdc31f4ea 100644
--- a/website/docs/gs/runtime_env.md
+++ b/website/docs/gs/runtime_env.md
@@ -1,24 +1,30 @@
---
-title: Runtime Environments
+title: Runtime environments
description: Describes the supported runtime environments.
-sidebar_position: 7
+sidebar_position: 5
slug: ./runtime-env
---
-The GATK-SV pipeline consists of _workflows_ and _reference data_ that
-orchestrates the analysis flow of input data. Hence, a successful
-execution requires running the _workflows_ on _reference_ and input data.
+The GATK-SV pipeline consists of workflows implemented in the Workflow Description Language
+([WDL](https://openwdl.org/)) and is built for use on the Google Cloud Platform ([GCP](https://cloud.google.com/)).
-:::info Currently supported backends: GCP
-GATK-SV has been tested only on the Google Cloud Platform (GCP);
-therefore, we are unable to provide specific guidance or support
-for other execution platforms including HPC clusters and AWS.
-:::
+### Terra (recommended)
+To facilitate easy-of-use, security, and collaboration, GATK-SV is available on the [Terra](https://app.terra.bio/)
+platform. Users should clone pre-configured Terra workspaces as a starting point for running GATK-SV:
-## Alternative backends
+- [Single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+- Joint calling workspace (TODO)
-Contributions from the community to improve portability between backends
-will be considered on a case-by-case-basis. We ask contributors to
+These workspaces are actively maintained by the development team and will be updated with critical fixes and major releases.
+
+### Cromwell (advanced)
+Advanced users and developers who wish to run GATK-SV on a dedicated Cromwell server using GCP as a backend should refer
+to the [Advanced Guides](/docs/category/advanced-guides) section.
+
+### Alternative backends (not supported)
+
+Use of other backends (e.g. AWS or on-prem HPC clusters) is not currently supported. However, contributions from the
+community to improve portability between backends will be considered on a case-by-case-basis. We ask contributors to
please adhere to the following guidelines when submitting issues and pull requests:
1. Code changes must be functionally equivalent on GCP backends, i.e. not result in changed output
diff --git a/website/docs/gs/sv_callers.md b/website/docs/gs/sv_callers.md
new file mode 100644
index 000000000..165bf87b3
--- /dev/null
+++ b/website/docs/gs/sv_callers.md
@@ -0,0 +1,28 @@
+---
+title: SV/CNV callers
+description: Summary of SV discovery tools
+sidebar_position: 3
+---
+
+GATK-SV uses an ensemble of SV discovery tools to produce a raw call set and then integrates, filters, refines,
+and annotates the calls from these tools to produce a final call set.
+
+The SV calling tools, sometimes referred to as "PE/SR" tools, include:
+- [Manta](https://github.com/Illumina/manta)
+- [Wham](https://github.com/zeeev/wham)
+- [Scramble](https://github.com/GeneDx/scramble)
+
+Depth-based calling of copy number variants (CNVs) is performed by two tools:
+- [GATK-gCNV](https://github.com/broadinstitute/gatk)
+- [cn.MOPS](https://bioconductor.org/packages/release/bioc/html/cn.mops.html)
+
+While it is possible to omit individual tools from the pipeline, it is strongly recommended to use the default
+configuration that runs all of them.
+
+:::note
+As of 2024, most published joint call sets generated with GATK-SV used the tool MELT, a state-of-the-art mobile element
+insertion (MEI) detector, instead of Scramble. Due to licensing restrictions, we cannot provide a public docker image
+or reference panel VCFs for this algorithm. The version of the pipeline configured in the Terra workspaces does not run
+MELT or include MELT calls for the reference panel. However, the Scramble tool has replaced MELT as an MEI caller and
+should provide similar performance.
+:::
diff --git a/website/docs/intro.md b/website/docs/intro.md
index 46d0da8a7..413f28245 100644
--- a/website/docs/intro.md
+++ b/website/docs/intro.md
@@ -4,29 +4,29 @@ description: GATK-SV
sidebar_position: 1
---
-GATK-SV is a comprehensive, cloud-based ensemble pipeline to capture and annotate all
-classes of structural variants (SV) from whole genome sequencing (WGS). It can detect
+GATK-SV is a comprehensive, cloud-based ensemble pipeline for discovering and annotating all
+classes of structural variants (SV) from whole genome sequencing (WGS) data. It can detect
deletions, duplications, multi-allelic copy number variants, balanced inversions,
insertions, translocations, and a diverse spectrum of complex SV. Briefly, GATK-SV
-maximizes the sensitivity of SV discovery by harmonizing output from five tools
-(Manta, Wham, cnMOPS, GATK-gCNV, MELT). In order to reduce false positives, raw SV
+maximizes the sensitivity of SV discovery by harmonizing output from five tools:
+Manta, Wham, Scramble, cnMOPS, and GATK-gCNV. To minimize false positives, raw SVs
are adjudicated and re-genotyped from read evidence considering all potential
-sequencing evidence including anomalous paired-end (PE) reads, split reads (SR) crossing
-a breakpoint, normalized read-depth (RD), and B-allele frequencies (BAF). It also
-fully resolves 11 classes of complex SV based on their breakpoint signature. GATK-SV
-has been designed to be deployed in the Google Cloud Platform via the cromwell
-execution engine, which allows massively parallel scaling. Further details about
-GATK--SV can be found in [Collins et al. 2020, Nature](https://www.nature.com/articles/s41586-020-2287-8).
+sequencing evidence including anomalous paired-end (PE) reads, split reads (SR),
+read-depth (RD), and B-allele frequencies (BAF). It also fully resolves 16 classes of complex
+SVs composed of multiple breakpoints. GATK-SV is intended for use on the [Terra](https://app.terra.bio/)
+ platform.
+### Methods
-A high-level description of GATK-SV is available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022487952155-Structural-variant-SV-discovery).
+Further details about GATK-SV methods can be found in [Collins et al. 2020](https://www.nature.com/articles/s41586-020-2287-8).
-### Citation
+### GATK Best Practices
-Please cite the following publication:
+Additional guidance on running GATK-SV is also available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022653744283-GATK-Best-Practices-for-Structural-Variation-Discovery-on-Single-Samples).
-- [Collins, Brand, et al. 2020. "A structural variation reference for medical and population genetics." Nature 581, 444-451.](https://doi.org/10.1038/s41586-020-2287-8)
+### Where to go from here
-Additional references:
+This documentation includes instructions for running the pipeline, technical implementation details, troubleshooting
+information, and guides for advanced users who wish to work with the source code or rebuild the project.
-- [Werling et al. 2018. "An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder." Nature genetics 50.5, 727-736.](https://doi.org/10.1038/s41588-018-0107-y)
+We recommend new users continue to the [Getting Started overview](/docs/gs/overview.md).
diff --git a/website/docs/modules/_category_.json b/website/docs/modules/_category_.json
index bb1a47f82..8f985b25a 100644
--- a/website/docs/modules/_category_.json
+++ b/website/docs/modules/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Modules",
- "position": 6,
+ "position": 5,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/modules/annotate_vcf.md b/website/docs/modules/annotate_vcf.md
index f4d965b22..1ec30cc47 100644
--- a/website/docs/modules/annotate_vcf.md
+++ b/website/docs/modules/annotate_vcf.md
@@ -1,23 +1,101 @@
---
-title: AnnotateVCF
-description: Annotate VCF (work in progress)
-sidebar_position: 13
+title: AnnotateVcf
+description: Annotate VCF
+sidebar_position: 20
slug: av
---
-Add annotations, such as the inferred function and
-allele frequencies of variants, to final VCF.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+Adds annotations, such as the inferred function and allele frequencies of variants, to a VCF.
Annotations methods include:
+* Functional annotation - The GATK tool [SVAnnotate](https://gatk.broadinstitute.org/hc/en-us/articles/13832752531355-SVAnnotate)
+ is used to annotate SVs with inferred functional consequence on protein-coding regions, regulatory regions such as
+ UTR and promoters, and other non-coding elements.
+* Allele Frequency (`AF`) annotation - annotate SVs with their allele frequencies across all samples, and samples of
+ specific sex, as well as specific subpopulations.
+* Allele Frequency annotation with external callset - annotate SVs with the allele frequencies of their overlapping SVs
+ in another callset, eg. the gnomAD-SV reference callset.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ fg: FilterGenotypes
+ avcf: AnnotateVcf
+ fg --> avcf
+
+ class avcf thisModule
+ class fg inModules
+```
+
+### Inputs
+
+#### `vcf`
+Any SV VCF. Running on the [genotype filtered VCF](./fg#filtered_vcf) is recommended.
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `protein_coding_gtf`
+Coding transcript definitions, see [here](/docs/resources#protein_coding_gtf).
+
+#### Optional `noncoding_bed`
+Non-coding reference intervals, see [here](//docs/resources#noncoding_bed).
+
+#### Optional `promoter_window`
+Promoter window size. See [here](https://gatk.broadinstitute.org/hc/en-us/articles/27007964610331-SVAnnotate#--promoter-window-length).
+
+#### Optional `max_breakend_as_cnv_length`
+Max size for treating `BND` records as CNVs. See [here](https://gatk.broadinstitute.org/hc/en-us/articles/27007964610331-SVAnnotate#--max-breakend-as-cnv-length).
+
+#### Optional `svannotate_additional_args`
+Additional arguments for [GATK-SVAnnotate](https://gatk.broadinstitute.org/hc/en-us/articles/27007964610331-SVAnnotate).
+
+#### Optional `sample_pop_assignments`
+Two-column file with sample ID & population assignment. "." for population will ignore the sample. If provided,
+annotates population-specific allele frequencies.
+
+#### Optional `sample_keep_list`
+If provided, subset samples to this list in the output VCF.
+
+#### Optional `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+If provided, sex-specific allele frequencies will be annotated.
+
+#### Optional `par_bed`
+Pseudo-autosomal region (PAR) bed file. If provided, variants overlapping PARs will be annotated with the `PAR` field.
+
+#### `sv_per_shard`
+Shard sized for parallel processing. Decreasing this may help if the workflow is running too slowly.
+
+#### Optional `external_af_ref_bed`
+Reference SV set (see [here](/docs/resources#external_af_ref_bed)). If provided, annotates variants with allele frequencies
+from the reference population.
+
+#### Optional `external_af_ref_prefix`
+External `AF` annotation prefix. Required if providing [external_af_ref_bed](#optional-external_af_ref_bed).
+
+#### Optional `external_af_population`
+Population names in the external SV reference set, e.g. "ALL", "AFR", "AMR", "EAS", "EUR". Required if providing
+[external_af_ref_bed](#optional-external_af_ref_bed) and must match the populations in the bed file.
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
-- Functional annotation - annotate SVs with inferred functional
- consequence on protein-coding regions, regulatory regions
- such as UTR and promoters, and other non-coding elements.
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
-- Allele Frequency annotation - annotate SVs with their allele
- frequencies across all samples, and samples of specific sex,
- as well as specific sub-populations.
+### Outputs
-- Allele Frequency annotation with external callset - annotate
- SVs with the allele frequencies of their overlapping SVs in
- another callset, eg. gnomad SV callset.
+#### `annotated_vcf`
+Output VCF.
diff --git a/website/docs/modules/apply_manual_filter.md b/website/docs/modules/apply_manual_filter.md
new file mode 100644
index 000000000..6c9bd7058
--- /dev/null
+++ b/website/docs/modules/apply_manual_filter.md
@@ -0,0 +1,57 @@
+---
+title: ApplyManualVariantFilter
+description: Complex SV genotyping
+sidebar_position: 16
+slug: amvf
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/ApplyManualVariantFilter.wdl)
+
+This module hard-filters variants (dropping records) using [bcftools](https://github.com/samtools/bcftools). While the
+workflow is general-purpose, we recommend running it with default parameters to eliminate major sources of false
+positive variants:
+
+1. Deletions called solely by `Wham`.
+2. SVA MEIs called by `Scramble` with the `HIGH_SR_BACKGROUND` flag.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ refcv: RefineComplexVariants
+ amvf: ApplyManualVariantFilter
+ svc: SVConcordance
+ refcv --> amvf
+ amvf --> svc
+
+ class amvf thisModule
+ class refcv inModules
+ class svc outModules
+```
+
+### Inputs
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `vcf`
+Any VCF. Running on the [cleaned VCF](cvcf#cleaned_vcf) is recommended.
+
+#### `filter_name`
+A name for the filter, used for output file naming. May be alphanumeric with underscores.
+
+#### `bcftools_filter`
+[Bcftools EXPRESSION](https://samtools.github.io/bcftools/bcftools.html#expressions) to use for filtering. Variants
+matching this expression will be **excluded**, i.e. with the `-e` argument.
+
+### Outputs
+
+#### `manual_filtered_vcf`
+Filtered VCF.
diff --git a/website/docs/modules/clean_vcf.md b/website/docs/modules/clean_vcf.md
new file mode 100644
index 000000000..66d56d0f5
--- /dev/null
+++ b/website/docs/modules/clean_vcf.md
@@ -0,0 +1,80 @@
+---
+title: CleanVcf
+description: VCF cleaning
+sidebar_position: 14
+slug: cvcf
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/CleanVcf.wdl)
+
+Performs various VCF clean-up steps including:
+
+- Adjusting genotypes on allosomal contigs
+- Collapsing overlapping CNVs into multi-allelic CNVs
+- Revising genotypes in overlapping CNVs
+- Removing redundant CNVs
+- Stitching large CNVs
+- VCF formatting clean-up
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ gcv: GenotypeComplexVariants
+ cvcf: CleanVcf
+ refcv: RefineComplexVariants
+
+ gcv --> cvcf
+ cvcf --> refcv
+
+ class cvcf thisModule
+ class gcv inModules
+ class refcv outModules
+```
+
+### Inputs
+
+#### `cohort_name`
+Cohort name. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `complex_genotype_vcfs`
+Array of contig-sharded VCFs containing genotyped complex variants, generated in [GenotypeComplexVariants](./gcv#complex_genotype_vcfs).
+
+#### `complex_resolve_bothside_pass_list`
+Array of variant lists with bothside SR support for all batches, generated in [ResolveComplexVariants](./rcv#complex_resolve_bothside_pass_list).
+
+#### `complex_resolve_background_fail_list`
+Array of variant lists with low SR signal-to-noise ratio for all batches, generated in [ResolveComplexVariants](./rcv#complex_resolve_background_fail_list).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+#### `max_shards_per_chrom_step1`, `min_records_per_shard_step1`, `samples_per_step2_shard`, `max_samples_per_shard_step3`, `clean_vcf1b_records_per_shard`, `clean_vcf5_records_per_shard`
+These parameters control parallelism in scattered tasks. Please examine the
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/CleanVcf.wdl) to see how each is used.
+
+#### Optional `outlier_samples_list`
+Text file of samples IDs to exclude when identifying multi-allelic CNVs. Most users do not need this feature unless
+excessive multi-allelic CNVs driven by low-quality samples are observed.
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `cleaned_vcf`
+Genome-wide VCF of output.
+
diff --git a/website/docs/modules/cluster_batch.md b/website/docs/modules/cluster_batch.md
index eb3435e24..0885d8aa7 100644
--- a/website/docs/modules/cluster_batch.md
+++ b/website/docs/modules/cluster_batch.md
@@ -5,19 +5,69 @@ sidebar_position: 5
slug: cb
---
-Clusters SV calls across a batch.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
-### Prerequisites
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/ClusterBatch.wdl)
-- GatherBatchEvidence
+Clusters SV calls across a batch. For each caller, redundant variants are merged across samples
+into representative variant records based on interval overlap criteria. Some variants will be hard-filtered
+if they overlap with predefined intervals known to pose challenges to SV and CNV callers (e.g. centromeres).
+[GATK-SVCluster](https://gatk.broadinstitute.org/hc/en-us/articles/27007962371099-SVCluster-BETA)
+is the primary tool used in for variant clustering.
+The following diagram illustrates the recommended invocation order:
-### Inputs
+```mermaid
-- Standardized call VCFs (GatherBatchEvidence)
-- Depth-only (DEL/DUP) calls (GatherBatchEvidence)
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
-### Outputs
+ gbe: GatherBatchEvidence
+ cb: ClusterBatch
+ gbm: GenerateBatchMetrics
+ jrc: JoinRawCalls
+
+ gbe --> cb
+ cb --> gbm
+ cb --> jrc
+
+ class cb thisModule
+ class gbe inModules
+ class gbm outModules
+ class jrc outModules
+```
-- Clustered SV VCFs
-- Clustered depth-only call VCF
+:::note
+[GenerateBatchMetrics](./gbm) is the primary downstream module in batch processing. [JoinRawCalls](./jrc) is
+required for genotype filtering but does not need to be run until later in the pipeline.
+:::
+
+## Inputs
+
+#### `batch`
+An identifier for the batch. Should match the name used in [GatherBatchEvidence](./gbe#batch).
+
+#### `*_vcf_tar`
+Standardized VCF tarballs from [GatherBatchEvidence](./gbe#std__vcf_tar)
+
+#### `del_bed`, `dup_bed`
+Merged CNV call files (`.bed.gz`) from [GatherBatchEvidence](./gbe#merged_dels-merged_dups)
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+#### Optional `N_IQR_cutoff_plotting`
+If provided, plot SV counts per sample. This number is used as the cutoff of interquartile range multiples for flagging
+outlier samples. Example value: 4.
+
+## Outputs
+
+#### `clustered_*_vcf`
+Clustered variants for each caller (`depth` corresponds to depth-based CNV callers `cnMOPS` and `GATK-gCNV`) in VCF format.
+
+#### Optional `clustered_sv_counts`, `clustered_sv_count_plots`, `clustered_outlier_samples_preview`, `clustered_outlier_samples_with_reason`, `clustered_num_outlier_samples`
+SV count QC tables and plots. Enable by providing [N_IQR_cutoff_plotting](#optional--n_iqr_cutoff_plotting)
diff --git a/website/docs/modules/combine_batches.md b/website/docs/modules/combine_batches.md
new file mode 100644
index 000000000..1fe6f5b14
--- /dev/null
+++ b/website/docs/modules/combine_batches.md
@@ -0,0 +1,92 @@
+---
+title: CombineBatches
+description: Cross-batch variant clustering
+sidebar_position: 11
+slug: cmb
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/CombineBatches.wdl)
+
+Merges variants across multiple batches. Variant merging uses similar methods and criteria as in [ClusterBatch](./cb),
+but in addition requires samples genotyped as non-reference to match sufficiently.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ rgc: RegenotypeCNVs
+ cb: CombineBatches
+ rcv: ResolveComplexVariants
+ rgc --> cb
+ cb --> rcv
+
+ class cb thisModule
+ class rgc inModules
+ class rcv outModules
+```
+
+### Inputs
+
+:::info
+All array inputs of batch data must match in order. For example, the order of the `batches` array should match that of
+`pesr_vcfs`, `depth_vcfs`, etc.
+:::
+
+#### `cohort_name`
+Cohort name. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `batches`
+Array of batch identifiers. Should match the name used in [GatherBatchEvidence](./gbe#batch). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### Optional `merge_vcfs`
+Default: `false`. If true, merge contig-sharded VCFs into one genome-wide VCF. This may be used for convenience but cannot be used with
+downstream workflows.
+
+#### `pesr_vcfs`
+Array of genotyped depth caller variants for all batches, generated in [GenotypeBatch](./gb#genotyped_depth_vcf).
+
+#### `depth_vcfs`
+Array of re-genotyped depth caller variants for all batches, generated in [RegenotypeCNVs](./rgcnvs#regenotyped_depth_vcfs).
+
+#### `raw_sr_bothside_pass_files`
+Array of variant lists with bothside SR support for all batches, generated in [GenotypeBatch](./gb#sr_bothside_pass).
+
+#### `raw_sr_background_fail_files`
+Array of variant lists with low SR signal-to-noise ratio for all batches, generated in [GenotypeBatch](./gb#sr_background_fail).
+
+#### `localize_shard_size`
+Shard size for parallel computations. Decreasing this parameter may help reduce run time.
+
+#### `min_sr_background_fail_batches`
+Threshold fraction of batches with high SR background for a given variant required in order to assign this
+`HIGH_SR_BACKGROUND` flag. Most users should leave this at the default value.
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `combined_vcfs`
+Array of contig-sharded VCFs of combined PE/SR and depth calls.
+
+#### `cluster_bothside_pass_lists`
+Array of contig-sharded bothside SR support variant lists.
+
+#### `cluster_background_fail_lists`
+Array of contig-sharded high SR background variant lists.
+
+#### `combine_batches_merged_vcf`
+Genome-wide VCF of combined PE/SR and depth calls. Only generated if using [merge_vcfs](#optional--merge_vcfs).
diff --git a/website/docs/modules/concordance.md b/website/docs/modules/concordance.md
new file mode 100644
index 000000000..16e8814ec
--- /dev/null
+++ b/website/docs/modules/concordance.md
@@ -0,0 +1,58 @@
+---
+title: SVConcordance
+description: Annotates concordance with raw calls
+sidebar_position: 18
+slug: svc
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/SVConcordance.wdl)
+
+Annotates variants with genotype concordance against another SV call set. This is a general-purpose workflow that can
+be applied to any pair of VCFs containing the same sample set. This is also a prerequisite step for genotype filtering
+in the recommended pipeline: genotypes are compared to calls emitted by raw callers, where low concordance can be indicative
+of poor quality variants. See
+[GATK-SVConcordance](https://gatk.broadinstitute.org/hc/en-us/articles/27007917991707-SVConcordance-BETA) for more
+information on methods.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ amvf: ApplyManualVariantFilter
+ jrc: JoinRawCalls
+ svc: SVConcordance
+ fg: FilterGenotypes
+ amvf --> svc
+ jrc --> svc
+ svc --> fg
+
+ class svc thisModule
+ class amvf inModules
+ class jrc inModules
+ class fg outModules
+```
+
+### Inputs
+
+#### `output_prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `eval_vcf`
+VCF to annotate. In the recommended pipeline, this is generated in [ApplyManualVariantFilter](./amvf).
+
+#### `truth_vcf`
+VCF to compare against. This should contain the same samples as `eval_vcf`. In the recommended pipeline, this is
+generated in [JoinRawCalls](./jrc).
+
+### Outputs
+
+#### `concordance_vcf`
+"Eval" VCF annotated with genotype concordance.
+
diff --git a/website/docs/modules/downstream_filter.md b/website/docs/modules/downstream_filter.md
deleted file mode 100644
index f06df455b..000000000
--- a/website/docs/modules/downstream_filter.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-title: Downstream Filtering
-description: Downstream filtering (work in progress)
-sidebar_position: 12
-slug: df
----
-
-Apply downstream filtering steps to the cleaned VCF to further
-control the false discovery rate; all steps are optional
-and users should decide based on the specific purpose of
-their projects.
-
-Filtering methods include:
-
-- minGQ - remove variants based on the genotype quality across
- populations. Note: Trio families are required to build the minGQ
- filtering model in this step. We provide tables pre-trained with
- the 1000 genomes samples at different FDR thresholds for projects
- that lack family structures, and they can be found at the paths below.
- These tables assume that GQ has a scale of [0,999], so they will
- not work with newer VCFs where GQ has a scale of [0,99].
-
- ```shell
- gs://gatk-sv-resources-public/hg38/v0/sv-resources/ref-panel/1KG/v2/mingq/1KGP_2504_and_698_with_GIAB.10perc_fdr.PCRMINUS.minGQ.filter_lookup_table.txt
- gs://gatk-sv-resources-public/hg38/v0/sv-resources/ref-panel/1KG/v2/mingq/1KGP_2504_and_698_with_GIAB.1perc_fdr.PCRMINUS.minGQ.filter_lookup_table.txt
- gs://gatk-sv-resources-public/hg38/v0/sv-resources/ref-panel/1KG/v2/mingq/1KGP_2504_and_698_with_GIAB.5perc_fdr.PCRMINUS.minGQ.filter_lookup_table.txt
- ```
-
-- BatchEffect - remove variants that show significant discrepancies
- in allele frequencies across batches
-
-- FilterOutlierSamplesPostMinGQ - remove outlier samples with unusually
- high or low number of SVs
-
-- FilterCleanupQualRecalibration - sanitize filter columns and
- recalibrate variant QUAL scores for easier interpretation
diff --git a/website/docs/modules/evidence_qc.md b/website/docs/modules/evidence_qc.md
index 5177636da..a48b3e0a8 100644
--- a/website/docs/modules/evidence_qc.md
+++ b/website/docs/modules/evidence_qc.md
@@ -7,31 +7,28 @@ slug: eqc
import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/EvidenceQC.wdl)
+
Runs ploidy estimation, dosage scoring, and optionally VCF QC.
The results from this module can be used for QC and batching.
For large cohorts, this workflow can be run on arbitrary cohort
-partitions of up to about 500 samples. Afterwards, we recommend
+partitions of up to about 500 samples. Afterward, we recommend
using the results to divide samples into smaller batches (~100-500 samples)
-with ~1:1 male:female ratio. Refer to the [Batching](/docs/run/joint#batching) section
+with ~1:1 male:female ratio. Refer to the [Batching](/docs/execution/joint#batching) section
for further guidance on creating batches.
We also recommend using sex assignments generated from the ploidy
estimates and incorporating them into the PED file, with sex = 0 for sex aneuploidies.
-The following diagram illustrates the upstream and downstream workflows of the `EvidenceQC` workflow
-in the recommended invocation order. You may refer to
-[this diagram](https://github.com/broadinstitute/gatk-sv/blob/main/terra_pipeline_diagram.jpg)
-for the overall recommended invocation order.
-
-
+
+### 10-GenotypeBatch
+
+Read the full GenotypeBatch documentation [here](/docs/modules/gb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `08-FilterBatchSamples`.
+
+### Steps 11-17
+
+Read the full documentation for [RegenotypeCNVs](/docs/modules/rgcnvs), [MakeCohortVcf](/docs/modules/cvcf) (which
+includes `CombineBatches`, `ResolveComplexVariants`, `GenotypeComplexVariants`, `CleanVcf`, `MainVcfQc`), and
+[AnnotateVcf](/docs/modules/av).
+* Use the same cohort `sample_set_set` you created and used for `09-MergeBatchSites`.
+
+### Additional notes
+
+- The VCF produced by `15-CleanVcf` (and annotated by `17-AnnotateVcf`) prioritizes sensitivity, but additional downstream
+filtration is recommended to improve specificity.
+
diff --git a/website/docs/execution/overview.md b/website/docs/execution/overview.md
new file mode 100644
index 000000000..fecfd3825
--- /dev/null
+++ b/website/docs/execution/overview.md
@@ -0,0 +1,10 @@
+---
+title: Overview
+description: Overview
+sidebar_position: 1
+slug: overview
+---
+
+This section provides technical documentation for the running single-sample and joint calling modes in Terra.
+
+Please review the [Getting Started](/docs/gs/overview) material before proceeding with this section.
diff --git a/website/docs/execution/single.md b/website/docs/execution/single.md
new file mode 100644
index 000000000..1132a2ac3
--- /dev/null
+++ b/website/docs/execution/single.md
@@ -0,0 +1,121 @@
+---
+title: Single-sample
+description: Run the pipeline on a single sample
+sidebar_position: 3
+slug: single
+---
+
+## Introduction
+
+**Extending SV detection to small datasets**
+
+The Single Sample pipeline is designed to facilitate running the methods developed for the cohort-mode GATK-SV pipeline on small data sets or in
+clinical contexts where batching large numbers of samples is not an option. To do so, it uses precomputed data, SV calls,
+and model parameters computed by the cohort pipeline on a reference panel composed of similar samples. The pipeline integrates this
+precomputed information with signals extracted from the input CRAM file to produce a call set similar in quality to results
+computed by the cohort pipeline in a computationally tractable and reproducible manner.
+
+
+## Terra workspace
+Users should clone the [Terra single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+which is configured with a demo sample to run on.
+Refer to the following sections for instructions on how to run the pipeline on your data using this workspace.
+
+## Data
+
+### Case sample
+
+The default workspace includes a NA12878 input WGS CRAM file configured in the workspace samples data table. This file is part of the
+high coverage (30X) [WGS data](https://www.internationalgenome.org/data-portal/data-collection/30x-grch38) for the 1000
+Genomes Project samples generated by the New York Genome Center and hosted in
+[AnVIL](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019).
+
+### Reference panel
+
+The reference panel configured in the workspace consists of data and calls computed from 156 publicly available samples
+chosen from the NYGC/AnVIL 1000 Genomes high coverage data linked above.
+
+Inputs to the pipeline for the reference panel include:
+- A precomputed SV callset VCF, and joint-called depth-based CNV call files
+- Raw calls for the reference panel samples from Manta and WHAM
+- Trained models for calling copy number variation in GATK gCNV case mode
+- Parameters learned by the cohort mode pipeline in training machine learning models on the reference panel samples.
+
+These resources are primarily configured in the "Workspace Data" for this workspace. However, several of the resources need
+to be passed to the workflow as large lists of files or strings. Due to Terra limitations on uploading data containing lists to the
+workspace data table, these resources are specified directly in the workflow configuration.
+
+### Reference resources
+
+The pipeline uses a number of resource and data files computed for the hg38 reference:
+- Reference sequences and indices
+- Genome annotation tracks such as segmental duplication and RepeatMasker tracks
+- Data used for annotation of called variants, including GenCode gene annotations and gnomAD site allele frequencies.
+
+## Single sample workflow
+
+### What does it do?
+
+The workflow `gatk-sv-single-sample` calls structural variations on a single input CRAM by running the GATK-SV Single Sample Pipeline end-to-end.
+
+#### What does it require as input?
+
+The workflow accepts a single CRAM or BAM file as input, configured in the following parameters:
+
+|Input Type|Input Name|Description|
+|---------|--------|--------------|
+|`String`|`sample_id`|Case sample identifier|
+|`File`|`bam_or_cram_file`|Path to the GCS location of the input CRAM or BAM file|
+|`String`|`batch`|Arbitrary name to be assigned to the run|
+
+#### Additional workspace-level inputs
+
+- Reference resources for hg38
+- Input data for the reference panel
+- The set of docker images used in the pipeline.
+
+Please contact GATK-SV developers if you are interested in customizing these
+inputs beyond their defaults.
+
+#### What does it return as output?
+
+|Output Type|Output Name|Description|
+|---------|--------|--------------|
+|`File`|`final_vcf`|SV VCF output for the pipeline. Includes all sites genotyped as variant in the case sample and genotypes for the reference panel. Sites are annotated with overlap of functional genome elements and allele frequencies of matching variants in gnomAD|
+|`File`|`final_vcf_idx`|Index file for `final_vcf`|
+|`File`|`final_bed`|Final output in BED format. Filter status, list of variant samples, and all VCF INFO fields are reported as additional columns.|
+|`File`|`metrics_file`|Metrics computed from the input data and intermediate and final VCFs. Includes metrics on the SV evidence, and on the number of variants called, broken down by type and size range.|
+|`File`|`qc_file`|Quality-control check file. This extracts several key metrics from the `metrics_file` and compares them to pre-specified threshold values. If any QC checks evaluate to FAIL, further diagnostics may be required.|
+|`File`|`ploidy_matrix`|Matrix of contig ploidy estimates computed by GATK gCNV.|
+|`File`|`ploidy_plots`|Plots of contig ploidy generated from `ploidy_matrix`|
+|`File`|`non_genotyped_unique_depth_calls`|This VCF file contains any depth based calls made in the case sample that did not pass genotyping checks and do not match a depth-based call from the reference panel. If very high sensitivity is desired, examine this file for additional large CNV calls.|
+|`File`|`non_genotyped_unique_depth_calls_idx`|Index file for `non_genotyped_unique_depth_calls`|
+|`File`|`pre_cleanup_vcf`|VCF output in a representation used internally in the pipeline. This file is less compliant with the VCF spec and is intended for debugging purposes.|
+|`File`|`pre_cleanup_vcf_idx`|Index file for `pre_cleanup_vcf`|
+
+#### Example time and cost run on sample data
+
+|Sample Name|Sample Size|Time|Cost $|
+|-----------|-----------|----|------|
+|NA12878|18.17 GiB|23hrs|~$7.34|
+
+#### To use this workflow on your own data
+
+If you would like to run this workflow on your own samples (which must be medium-to-high coverage WGS data):
+
+- Clone the [workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample) into a Terra project you have access to
+- In the cloned workspace, upload rows to the Sample and (optionally) the Participant Data Table that describe your samples.
+ Ensure that the rows you add to the Sample table contain the columns `sample_id` and `bam_or_cram_file` are populated appropriately.
+- There is no need to modify values in the workspace data or method configuration. If you are interested in modifying the reference
+ genome resources or reference panel, please contact the GATK team for support as listed below.
+- Launch the workflow from the "Workflows" tab, selecting your samples as the inputs.
+
+#### Quality control
+
+Please check the `qc_file` output to screen for data quality issues. This file provides a table of quality metrics and
+suggested acceptable ranges.
+
+:::warning
+If a metric exceeds the recommended range, all variants will be automatically flagged with
+a non-passing `FILTER` status in the output VCF.
+:::
\ No newline at end of file
diff --git a/website/docs/gs/_category_.json b/website/docs/gs/_category_.json
index 6b9e1272a..fe28ad97e 100644
--- a/website/docs/gs/_category_.json
+++ b/website/docs/gs/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Getting Started",
- "position": 2,
+ "position": 3,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/gs/calling_modes.md b/website/docs/gs/calling_modes.md
new file mode 100644
index 000000000..dcafa5634
--- /dev/null
+++ b/website/docs/gs/calling_modes.md
@@ -0,0 +1,41 @@
+---
+title: Calling modes
+description: Description of single-sample and joint calling
+sidebar_position: 1
+---
+
+# Calling modes
+
+GATK-SV offers two different modes for SV calling. Users should carefully review the following sections to determine
+which mode is appropriate for their use case.
+
+## Single-sample mode
+
+GATK-SV can perform SV calling on individual samples. In this mode, a sample is jointly called against a fixed reference
+panel of [156 high-quality samples from the 1000 Genomes Project](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019). Single-sample mode is a good option for the following
+use cases:
+
+- Studies involving fewer 100 samples
+- Studies with rolling data delivery, i.e. in small batches over time
+
+Users should also consider that the single-sample mode is provided as a single workflow and is therefore considerably
+simpler to run than joint calling. However, it also has higher compute costs on a per-sample basis and will not be as sensitive
+as joint calling with larger cohorts.
+
+## Joint calling mode
+
+GATK-SV can also perform joint calling on a set of samples. Users may opt for this mode in the following use cases:
+
+- Studies involving at least 100 samples
+- When maximum sensitivity is desired
+- Data sets that are technically heterogeneous, i.e. with strong batch effects, or are very different from the single-sample mode reference panel
+
+Joint calling has the advantage of increasing SV discovery sensitivity and providing allele frequency estimates, and there are
+some features, such as genotype recalibration and filtering and in-depth QC plotting, that are only available in joint calling mode.
+However, this pipeline is considerably more complex to execute than the single-sample mode, requiring sample batching and the execution of
+several individual modules.
+
+## Related content
+
+More information on single-sample and joint calling can be found in the [Execution](/docs/execution/overview) section.
+
diff --git a/website/docs/gs/dockers.md b/website/docs/gs/dockers.md
new file mode 100644
index 000000000..7a4e0f658
--- /dev/null
+++ b/website/docs/gs/dockers.md
@@ -0,0 +1,73 @@
+---
+title: Docker images
+description: Docker images
+sidebar_position: 4
+---
+
+GATK-SV utilizes a set of [Docker](https://www.docker.com/) images for execution within containerized environments.
+
+### Publishing and availability
+
+Dockers are automatically built and pushed to the `us.gcr.io/broad-dsde-methods/gatk-sv` repository under two different conditions:
+1. **Release**: upon releasing a new version of GATK-SV. These Dockers are made permanently available.
+2. **Commit**: upon merging a new commit to the development branch. These Dockers are ephemeral and may be periodically
+deleted. Any users needing to preserve access to these Docker images should copy them to their own repository. Also
+note that these images are built on an "as-needed" basis, meaning an image is only updated if it has been changed
+in any way by the commit.
+
+The full set of current images are automatically published and pushed to a public
+[Google Artifact Registry](https://cloud.google.com/artifact-registry/docs) and listed in the
+[dockers.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers.json)
+file.
+
+:::info
+Microsoft Azure mirrors of all images can be found in
+[dockers_azure.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers_azure.json),
+but these are not currently available for public use.
+:::
+
+### Regions (IMPORTANT)
+
+Users using Google Compute VMs outside of the `us-central1` region, i.e. in other NA regions or continents, must copy all
+docker images to a repository hosted in their region. If using a Terra workspace, the region is listed under `Cloud
+Information`:`Location` in the workspace dashboard. Please see
+[this article](https://support.terra.bio/hc/en-us/articles/4408985788187-How-to-configure-Google-Artifact-Registry-to-prevent-data-transfer-egress-charges)
+for more information.
+
+:::warning
+Failure to localize Docker images to your region will incur significant egress costs.
+:::
+
+### Versioning
+
+All Docker images are tagged with a date and version number that must be run with the corresponding version of the
+WDLs. The Docker images built with a particular version can be determined from the `dockers.json` file by checking out
+the commit or release of interest and examining `dockers.json`, e.g.
+[v0.29-beta](https://github.com/broadinstitute/gatk-sv/blob/v0.29-beta/inputs/values/dockers.json).
+
+Note that a given commit may contain a mixture of Docker image version tags if only a subset of images has actually
+been updated, reflecting the "as-needed" rebuild policy described above.
+
+All Terra workspace releases have the WDLs and Docker images synchronized.
+
+:::info
+Cloning a Terra workspace copies a snapshot of the current workspace. Any future updates to the original workspace
+will not be propagated to the clone.
+:::
+
+:::warning
+We strongly recommend using a single version of GATK-SV for all projects. Switching versions may lead to batch effects
+and/or compromise call set fidelity.
+:::
+
+
+### Workflow inputs
+
+All GATK-SV WDLs utilize a subset of the Docker images, which are specified as inputs. For example,
+the `sv_pipeline` docker is supplied to the input `sv_pipeline_docker` in many WDLs.
+
+
+### Builds
+
+An in-depth guide to the GATK-SV Docker system and building Docker images is available in the
+[Advanced section](/docs/category/docker-builds).
diff --git a/website/docs/gs/input_files.md b/website/docs/gs/input_files.md
index 38e143869..aafd031f1 100644
--- a/website/docs/gs/input_files.md
+++ b/website/docs/gs/input_files.md
@@ -1,62 +1,72 @@
---
-title: Input Data
+title: Input data
description: Supported input and reference data.
-sidebar_position: 5
+sidebar_position: 2
slug: ./inputs
---
GATK-SV requires the following input data:
-- Illumina short-read whole-genome CRAMs or BAMs, aligned to hg38 with [bwa-mem](https://github.com/lh3/bwa).
- BAMs must also be indexed.
+1. Sequencing alignments in BAM or CRAM format that are:
+ - Short-read, paired-end Illumina (e.g. Novaseq)
+ - Deep whole-genome coverage (~30x); RNA-seq and targeted (exome) libraries are not supported
+ - Indexed (have a companion `.bai` or `.crai` file)
+ - Aligned to
+ [hg38](https://gatk.broadinstitute.org/hc/en-us/articles/360035890951-Human-genome-reference-builds-GRCh38-or-hg38-b37-hg19)
+ with either [GATK Best Practices](https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows)
+ and [bwa-mem](https://github.com/lh3/bwa),
+ or [Illumina DRAGEN](https://www.illumina.com/products/by-type/informatics-products/dragen-secondary-analysis.html) v3.4.12 or v3.7.8
+2. (Joint calling mode only) Family structure definitions file in [PED format](/docs/gs/inputs#ped-format). This file is required even if your dataset does not contain related individuals.
-- Family structure definitions file in
- [PED format](/docs/gs/inputs#ped-format).
-
-### PED file format {#ped-format}
-The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
-* The file must be tab-delimited.
-* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
-* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
-* Missing parental IDs should be entered as 0.
-* Header lines are allowed if they begin with a # character.
-To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
+Note that the supported alignment pipeline versions have been extensively tested for robustness and accuracy. While other
+versions of DRAGEN may work as well, they have not been validated with GATK-SV. We do not recommend mixing aligners within call sets.
### Sample Exclusion
We recommend filtering out samples with a high percentage
-of improperly paired reads (>10% or an outlier for your data)
+of improperly paired or chimeric reads
as technical outliers prior to running [GatherSampleEvidence](/docs/modules/gse).
-A high percentage of improperly paired reads may indicate issues
-with library prep, degradation, or contamination. Artifactual
-improperly paired reads could cause incorrect SV calls, and
-these samples have been observed to have longer runtimes and
-higher compute costs for [GatherSampleEvidence](/docs/modules/gse).
+Samples with high rates of anomalous reads may indicate issues
+with library preparation, degradation, or contamination and can lead to poor variant set quality.
+Samples failing these criteria often require longer run times and higher compute costs.
-### Sample ID requirements {#sampleids}
-#### Sample IDs must
+### Sample IDs {#sampleids}
+GATK-SV imposes certain restrictions on sample names (IDs) in order to avoid certain parsing errors (e.g. with the
+use of the `grep` command). While future releases will obviate some of these restrictions, users must modify
+their sample IDs according to the following requirements.
+
+#### Sample IDs must:
- Be unique within the cohort
- Contain only alphanumeric characters and underscores (no dashes, whitespace, or special characters)
-#### Sample IDs should not
-
-- Contain only numeric characters
+#### Sample IDs should not:
+- Contain only numeric characters, e.g. `10004928`
- Be a substring of another sample ID in the same cohort
- Contain any of the following substrings: `chr`, `name`, `DEL`, `DUP`, `CPX`, `CHROM`
-The same requirements apply to family IDs in the PED file,
-as well as batch IDs and the cohort ID provided as workflow inputs.
+The same requirements apply to family IDs in the PED file, as well as batch IDs and the cohort ID provided as workflow inputs.
-Sample IDs are provided to GatherSampleEvidence directly and
-need not match sample names from the BAM/CRAM headers.
-`GetSampleID.wdl` can be used to fetch BAM sample IDs and
-also generates a set of alternate IDs that are considered
-safe for this pipeline; alternatively, [this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
+Users should set sample IDs in [GatherSampleEvidence](/docs/modules/gse) with the `sample_id` input, which needs not match
+the sample name defined in the BAM/CRAM header. `GetSampleID.wdl` can be used to fetch BAM sample IDs and also generates a set
+of alternate IDs that are considered safe for this pipeline. Alternatively,
+[this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
transforms a list of sample IDs to fit these requirements.
-Currently, sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe).
-The following inputs will need to be updated with the transformed sample IDs:
+Sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe). To do so, set the parameter
+`rename_samples = True` and provide updated sample IDs via the `samples` parameter.
+
+Note that following inputs will need to be updated with the transformed sample IDs:
- Sample ID list for [GatherSampleEvidence](/docs/modules/gse) or [GatherBatchEvidence](/docs/modules/gbe)
- PED file
+
+
+### PED file format {#ped-format}
+The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
+* The file must be tab-delimited.
+* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
+* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
+* Missing parental IDs should be entered as 0.
+* Header lines are allowed if they begin with a # character.
+* To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
diff --git a/website/docs/gs/overview.md b/website/docs/gs/overview.md
index 5b147c973..f4493759c 100644
--- a/website/docs/gs/overview.md
+++ b/website/docs/gs/overview.md
@@ -6,9 +6,11 @@ sidebar_position: 0
# Overview
-GATK-SV is a highly-scalable cloud-native pipeline for structural variation discovery
-on Illumina short-read whole-genome sequencing (WGS) data.
-The pipeline genotypes structural variations using Docker-based tools, modularized in
-different components, and orchestrated using Cromwell.
+This section provides guidance on how GATK-SV can be used, restrictions on input data, and where to get started
+with generating call sets.
-Please refer to the documentation on [this page](/docs/run/overview) for running the pipeline.
+We highly recommend that users new to SV calling also review the [Best Practices Guide](/docs/best_practices).
+
+Most users should use the Terra platform for running GATK-SV and, after reading this section,
+should refer to the documentation in the [Execution section](/docs/execution/overview) for instructions on
+how to run their own data.
diff --git a/website/docs/gs/runtime_env.md b/website/docs/gs/runtime_env.md
index 65aee6032..cdc31f4ea 100644
--- a/website/docs/gs/runtime_env.md
+++ b/website/docs/gs/runtime_env.md
@@ -1,24 +1,30 @@
---
-title: Runtime Environments
+title: Runtime environments
description: Describes the supported runtime environments.
-sidebar_position: 7
+sidebar_position: 5
slug: ./runtime-env
---
-The GATK-SV pipeline consists of _workflows_ and _reference data_ that
-orchestrates the analysis flow of input data. Hence, a successful
-execution requires running the _workflows_ on _reference_ and input data.
+The GATK-SV pipeline consists of workflows implemented in the Workflow Description Language
+([WDL](https://openwdl.org/)) and is built for use on the Google Cloud Platform ([GCP](https://cloud.google.com/)).
-:::info Currently supported backends: GCP
-GATK-SV has been tested only on the Google Cloud Platform (GCP);
-therefore, we are unable to provide specific guidance or support
-for other execution platforms including HPC clusters and AWS.
-:::
+### Terra (recommended)
+To facilitate easy-of-use, security, and collaboration, GATK-SV is available on the [Terra](https://app.terra.bio/)
+platform. Users should clone pre-configured Terra workspaces as a starting point for running GATK-SV:
-## Alternative backends
+- [Single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+- Joint calling workspace (TODO)
-Contributions from the community to improve portability between backends
-will be considered on a case-by-case-basis. We ask contributors to
+These workspaces are actively maintained by the development team and will be updated with critical fixes and major releases.
+
+### Cromwell (advanced)
+Advanced users and developers who wish to run GATK-SV on a dedicated Cromwell server using GCP as a backend should refer
+to the [Advanced Guides](/docs/category/advanced-guides) section.
+
+### Alternative backends (not supported)
+
+Use of other backends (e.g. AWS or on-prem HPC clusters) is not currently supported. However, contributions from the
+community to improve portability between backends will be considered on a case-by-case-basis. We ask contributors to
please adhere to the following guidelines when submitting issues and pull requests:
1. Code changes must be functionally equivalent on GCP backends, i.e. not result in changed output
diff --git a/website/docs/gs/sv_callers.md b/website/docs/gs/sv_callers.md
new file mode 100644
index 000000000..165bf87b3
--- /dev/null
+++ b/website/docs/gs/sv_callers.md
@@ -0,0 +1,28 @@
+---
+title: SV/CNV callers
+description: Summary of SV discovery tools
+sidebar_position: 3
+---
+
+GATK-SV uses an ensemble of SV discovery tools to produce a raw call set and then integrates, filters, refines,
+and annotates the calls from these tools to produce a final call set.
+
+The SV calling tools, sometimes referred to as "PE/SR" tools, include:
+- [Manta](https://github.com/Illumina/manta)
+- [Wham](https://github.com/zeeev/wham)
+- [Scramble](https://github.com/GeneDx/scramble)
+
+Depth-based calling of copy number variants (CNVs) is performed by two tools:
+- [GATK-gCNV](https://github.com/broadinstitute/gatk)
+- [cn.MOPS](https://bioconductor.org/packages/release/bioc/html/cn.mops.html)
+
+While it is possible to omit individual tools from the pipeline, it is strongly recommended to use the default
+configuration that runs all of them.
+
+:::note
+As of 2024, most published joint call sets generated with GATK-SV used the tool MELT, a state-of-the-art mobile element
+insertion (MEI) detector, instead of Scramble. Due to licensing restrictions, we cannot provide a public docker image
+or reference panel VCFs for this algorithm. The version of the pipeline configured in the Terra workspaces does not run
+MELT or include MELT calls for the reference panel. However, the Scramble tool has replaced MELT as an MEI caller and
+should provide similar performance.
+:::
diff --git a/website/docs/intro.md b/website/docs/intro.md
index 46d0da8a7..413f28245 100644
--- a/website/docs/intro.md
+++ b/website/docs/intro.md
@@ -4,29 +4,29 @@ description: GATK-SV
sidebar_position: 1
---
-GATK-SV is a comprehensive, cloud-based ensemble pipeline to capture and annotate all
-classes of structural variants (SV) from whole genome sequencing (WGS). It can detect
+GATK-SV is a comprehensive, cloud-based ensemble pipeline for discovering and annotating all
+classes of structural variants (SV) from whole genome sequencing (WGS) data. It can detect
deletions, duplications, multi-allelic copy number variants, balanced inversions,
insertions, translocations, and a diverse spectrum of complex SV. Briefly, GATK-SV
-maximizes the sensitivity of SV discovery by harmonizing output from five tools
-(Manta, Wham, cnMOPS, GATK-gCNV, MELT). In order to reduce false positives, raw SV
+maximizes the sensitivity of SV discovery by harmonizing output from five tools:
+Manta, Wham, Scramble, cnMOPS, and GATK-gCNV. To minimize false positives, raw SVs
are adjudicated and re-genotyped from read evidence considering all potential
-sequencing evidence including anomalous paired-end (PE) reads, split reads (SR) crossing
-a breakpoint, normalized read-depth (RD), and B-allele frequencies (BAF). It also
-fully resolves 11 classes of complex SV based on their breakpoint signature. GATK-SV
-has been designed to be deployed in the Google Cloud Platform via the cromwell
-execution engine, which allows massively parallel scaling. Further details about
-GATK--SV can be found in [Collins et al. 2020, Nature](https://www.nature.com/articles/s41586-020-2287-8).
+sequencing evidence including anomalous paired-end (PE) reads, split reads (SR),
+read-depth (RD), and B-allele frequencies (BAF). It also fully resolves 16 classes of complex
+SVs composed of multiple breakpoints. GATK-SV is intended for use on the [Terra](https://app.terra.bio/)
+ platform.
+### Methods
-A high-level description of GATK-SV is available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022487952155-Structural-variant-SV-discovery).
+Further details about GATK-SV methods can be found in [Collins et al. 2020](https://www.nature.com/articles/s41586-020-2287-8).
-### Citation
+### GATK Best Practices
-Please cite the following publication:
+Additional guidance on running GATK-SV is also available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022653744283-GATK-Best-Practices-for-Structural-Variation-Discovery-on-Single-Samples).
-- [Collins, Brand, et al. 2020. "A structural variation reference for medical and population genetics." Nature 581, 444-451.](https://doi.org/10.1038/s41586-020-2287-8)
+### Where to go from here
-Additional references:
+This documentation includes instructions for running the pipeline, technical implementation details, troubleshooting
+information, and guides for advanced users who wish to work with the source code or rebuild the project.
-- [Werling et al. 2018. "An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder." Nature genetics 50.5, 727-736.](https://doi.org/10.1038/s41588-018-0107-y)
+We recommend new users continue to the [Getting Started overview](/docs/gs/overview.md).
diff --git a/website/docs/modules/_category_.json b/website/docs/modules/_category_.json
index bb1a47f82..8f985b25a 100644
--- a/website/docs/modules/_category_.json
+++ b/website/docs/modules/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Modules",
- "position": 6,
+ "position": 5,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/modules/annotate_vcf.md b/website/docs/modules/annotate_vcf.md
index f4d965b22..1ec30cc47 100644
--- a/website/docs/modules/annotate_vcf.md
+++ b/website/docs/modules/annotate_vcf.md
@@ -1,23 +1,101 @@
---
-title: AnnotateVCF
-description: Annotate VCF (work in progress)
-sidebar_position: 13
+title: AnnotateVcf
+description: Annotate VCF
+sidebar_position: 20
slug: av
---
-Add annotations, such as the inferred function and
-allele frequencies of variants, to final VCF.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+Adds annotations, such as the inferred function and allele frequencies of variants, to a VCF.
Annotations methods include:
+* Functional annotation - The GATK tool [SVAnnotate](https://gatk.broadinstitute.org/hc/en-us/articles/13832752531355-SVAnnotate)
+ is used to annotate SVs with inferred functional consequence on protein-coding regions, regulatory regions such as
+ UTR and promoters, and other non-coding elements.
+* Allele Frequency (`AF`) annotation - annotate SVs with their allele frequencies across all samples, and samples of
+ specific sex, as well as specific subpopulations.
+* Allele Frequency annotation with external callset - annotate SVs with the allele frequencies of their overlapping SVs
+ in another callset, eg. the gnomAD-SV reference callset.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ fg: FilterGenotypes
+ avcf: AnnotateVcf
+ fg --> avcf
+
+ class avcf thisModule
+ class fg inModules
+```
+
+### Inputs
+
+#### `vcf`
+Any SV VCF. Running on the [genotype filtered VCF](./fg#filtered_vcf) is recommended.
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `protein_coding_gtf`
+Coding transcript definitions, see [here](/docs/resources#protein_coding_gtf).
+
+#### Optional `noncoding_bed`
+Non-coding reference intervals, see [here](//docs/resources#noncoding_bed).
+
+#### Optional `promoter_window`
+Promoter window size. See [here](https://gatk.broadinstitute.org/hc/en-us/articles/27007964610331-SVAnnotate#--promoter-window-length).
+
+#### Optional `max_breakend_as_cnv_length`
+Max size for treating `BND` records as CNVs. See [here](https://gatk.broadinstitute.org/hc/en-us/articles/27007964610331-SVAnnotate#--max-breakend-as-cnv-length).
+
+#### Optional `svannotate_additional_args`
+Additional arguments for [GATK-SVAnnotate](https://gatk.broadinstitute.org/hc/en-us/articles/27007964610331-SVAnnotate).
+
+#### Optional `sample_pop_assignments`
+Two-column file with sample ID & population assignment. "." for population will ignore the sample. If provided,
+annotates population-specific allele frequencies.
+
+#### Optional `sample_keep_list`
+If provided, subset samples to this list in the output VCF.
+
+#### Optional `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+If provided, sex-specific allele frequencies will be annotated.
+
+#### Optional `par_bed`
+Pseudo-autosomal region (PAR) bed file. If provided, variants overlapping PARs will be annotated with the `PAR` field.
+
+#### `sv_per_shard`
+Shard sized for parallel processing. Decreasing this may help if the workflow is running too slowly.
+
+#### Optional `external_af_ref_bed`
+Reference SV set (see [here](/docs/resources#external_af_ref_bed)). If provided, annotates variants with allele frequencies
+from the reference population.
+
+#### Optional `external_af_ref_prefix`
+External `AF` annotation prefix. Required if providing [external_af_ref_bed](#optional-external_af_ref_bed).
+
+#### Optional `external_af_population`
+Population names in the external SV reference set, e.g. "ALL", "AFR", "AMR", "EAS", "EUR". Required if providing
+[external_af_ref_bed](#optional-external_af_ref_bed) and must match the populations in the bed file.
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
-- Functional annotation - annotate SVs with inferred functional
- consequence on protein-coding regions, regulatory regions
- such as UTR and promoters, and other non-coding elements.
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
-- Allele Frequency annotation - annotate SVs with their allele
- frequencies across all samples, and samples of specific sex,
- as well as specific sub-populations.
+### Outputs
-- Allele Frequency annotation with external callset - annotate
- SVs with the allele frequencies of their overlapping SVs in
- another callset, eg. gnomad SV callset.
+#### `annotated_vcf`
+Output VCF.
diff --git a/website/docs/modules/apply_manual_filter.md b/website/docs/modules/apply_manual_filter.md
new file mode 100644
index 000000000..6c9bd7058
--- /dev/null
+++ b/website/docs/modules/apply_manual_filter.md
@@ -0,0 +1,57 @@
+---
+title: ApplyManualVariantFilter
+description: Complex SV genotyping
+sidebar_position: 16
+slug: amvf
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/ApplyManualVariantFilter.wdl)
+
+This module hard-filters variants (dropping records) using [bcftools](https://github.com/samtools/bcftools). While the
+workflow is general-purpose, we recommend running it with default parameters to eliminate major sources of false
+positive variants:
+
+1. Deletions called solely by `Wham`.
+2. SVA MEIs called by `Scramble` with the `HIGH_SR_BACKGROUND` flag.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ refcv: RefineComplexVariants
+ amvf: ApplyManualVariantFilter
+ svc: SVConcordance
+ refcv --> amvf
+ amvf --> svc
+
+ class amvf thisModule
+ class refcv inModules
+ class svc outModules
+```
+
+### Inputs
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `vcf`
+Any VCF. Running on the [cleaned VCF](cvcf#cleaned_vcf) is recommended.
+
+#### `filter_name`
+A name for the filter, used for output file naming. May be alphanumeric with underscores.
+
+#### `bcftools_filter`
+[Bcftools EXPRESSION](https://samtools.github.io/bcftools/bcftools.html#expressions) to use for filtering. Variants
+matching this expression will be **excluded**, i.e. with the `-e` argument.
+
+### Outputs
+
+#### `manual_filtered_vcf`
+Filtered VCF.
diff --git a/website/docs/modules/clean_vcf.md b/website/docs/modules/clean_vcf.md
new file mode 100644
index 000000000..66d56d0f5
--- /dev/null
+++ b/website/docs/modules/clean_vcf.md
@@ -0,0 +1,80 @@
+---
+title: CleanVcf
+description: VCF cleaning
+sidebar_position: 14
+slug: cvcf
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/CleanVcf.wdl)
+
+Performs various VCF clean-up steps including:
+
+- Adjusting genotypes on allosomal contigs
+- Collapsing overlapping CNVs into multi-allelic CNVs
+- Revising genotypes in overlapping CNVs
+- Removing redundant CNVs
+- Stitching large CNVs
+- VCF formatting clean-up
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ gcv: GenotypeComplexVariants
+ cvcf: CleanVcf
+ refcv: RefineComplexVariants
+
+ gcv --> cvcf
+ cvcf --> refcv
+
+ class cvcf thisModule
+ class gcv inModules
+ class refcv outModules
+```
+
+### Inputs
+
+#### `cohort_name`
+Cohort name. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `complex_genotype_vcfs`
+Array of contig-sharded VCFs containing genotyped complex variants, generated in [GenotypeComplexVariants](./gcv#complex_genotype_vcfs).
+
+#### `complex_resolve_bothside_pass_list`
+Array of variant lists with bothside SR support for all batches, generated in [ResolveComplexVariants](./rcv#complex_resolve_bothside_pass_list).
+
+#### `complex_resolve_background_fail_list`
+Array of variant lists with low SR signal-to-noise ratio for all batches, generated in [ResolveComplexVariants](./rcv#complex_resolve_background_fail_list).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+#### `max_shards_per_chrom_step1`, `min_records_per_shard_step1`, `samples_per_step2_shard`, `max_samples_per_shard_step3`, `clean_vcf1b_records_per_shard`, `clean_vcf5_records_per_shard`
+These parameters control parallelism in scattered tasks. Please examine the
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/CleanVcf.wdl) to see how each is used.
+
+#### Optional `outlier_samples_list`
+Text file of samples IDs to exclude when identifying multi-allelic CNVs. Most users do not need this feature unless
+excessive multi-allelic CNVs driven by low-quality samples are observed.
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `cleaned_vcf`
+Genome-wide VCF of output.
+
diff --git a/website/docs/modules/cluster_batch.md b/website/docs/modules/cluster_batch.md
index eb3435e24..0885d8aa7 100644
--- a/website/docs/modules/cluster_batch.md
+++ b/website/docs/modules/cluster_batch.md
@@ -5,19 +5,69 @@ sidebar_position: 5
slug: cb
---
-Clusters SV calls across a batch.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
-### Prerequisites
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/ClusterBatch.wdl)
-- GatherBatchEvidence
+Clusters SV calls across a batch. For each caller, redundant variants are merged across samples
+into representative variant records based on interval overlap criteria. Some variants will be hard-filtered
+if they overlap with predefined intervals known to pose challenges to SV and CNV callers (e.g. centromeres).
+[GATK-SVCluster](https://gatk.broadinstitute.org/hc/en-us/articles/27007962371099-SVCluster-BETA)
+is the primary tool used in for variant clustering.
+The following diagram illustrates the recommended invocation order:
-### Inputs
+```mermaid
-- Standardized call VCFs (GatherBatchEvidence)
-- Depth-only (DEL/DUP) calls (GatherBatchEvidence)
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
-### Outputs
+ gbe: GatherBatchEvidence
+ cb: ClusterBatch
+ gbm: GenerateBatchMetrics
+ jrc: JoinRawCalls
+
+ gbe --> cb
+ cb --> gbm
+ cb --> jrc
+
+ class cb thisModule
+ class gbe inModules
+ class gbm outModules
+ class jrc outModules
+```
-- Clustered SV VCFs
-- Clustered depth-only call VCF
+:::note
+[GenerateBatchMetrics](./gbm) is the primary downstream module in batch processing. [JoinRawCalls](./jrc) is
+required for genotype filtering but does not need to be run until later in the pipeline.
+:::
+
+## Inputs
+
+#### `batch`
+An identifier for the batch. Should match the name used in [GatherBatchEvidence](./gbe#batch).
+
+#### `*_vcf_tar`
+Standardized VCF tarballs from [GatherBatchEvidence](./gbe#std__vcf_tar)
+
+#### `del_bed`, `dup_bed`
+Merged CNV call files (`.bed.gz`) from [GatherBatchEvidence](./gbe#merged_dels-merged_dups)
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+#### Optional `N_IQR_cutoff_plotting`
+If provided, plot SV counts per sample. This number is used as the cutoff of interquartile range multiples for flagging
+outlier samples. Example value: 4.
+
+## Outputs
+
+#### `clustered_*_vcf`
+Clustered variants for each caller (`depth` corresponds to depth-based CNV callers `cnMOPS` and `GATK-gCNV`) in VCF format.
+
+#### Optional `clustered_sv_counts`, `clustered_sv_count_plots`, `clustered_outlier_samples_preview`, `clustered_outlier_samples_with_reason`, `clustered_num_outlier_samples`
+SV count QC tables and plots. Enable by providing [N_IQR_cutoff_plotting](#optional--n_iqr_cutoff_plotting)
diff --git a/website/docs/modules/combine_batches.md b/website/docs/modules/combine_batches.md
new file mode 100644
index 000000000..1fe6f5b14
--- /dev/null
+++ b/website/docs/modules/combine_batches.md
@@ -0,0 +1,92 @@
+---
+title: CombineBatches
+description: Cross-batch variant clustering
+sidebar_position: 11
+slug: cmb
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/CombineBatches.wdl)
+
+Merges variants across multiple batches. Variant merging uses similar methods and criteria as in [ClusterBatch](./cb),
+but in addition requires samples genotyped as non-reference to match sufficiently.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ rgc: RegenotypeCNVs
+ cb: CombineBatches
+ rcv: ResolveComplexVariants
+ rgc --> cb
+ cb --> rcv
+
+ class cb thisModule
+ class rgc inModules
+ class rcv outModules
+```
+
+### Inputs
+
+:::info
+All array inputs of batch data must match in order. For example, the order of the `batches` array should match that of
+`pesr_vcfs`, `depth_vcfs`, etc.
+:::
+
+#### `cohort_name`
+Cohort name. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `batches`
+Array of batch identifiers. Should match the name used in [GatherBatchEvidence](./gbe#batch). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### Optional `merge_vcfs`
+Default: `false`. If true, merge contig-sharded VCFs into one genome-wide VCF. This may be used for convenience but cannot be used with
+downstream workflows.
+
+#### `pesr_vcfs`
+Array of genotyped depth caller variants for all batches, generated in [GenotypeBatch](./gb#genotyped_depth_vcf).
+
+#### `depth_vcfs`
+Array of re-genotyped depth caller variants for all batches, generated in [RegenotypeCNVs](./rgcnvs#regenotyped_depth_vcfs).
+
+#### `raw_sr_bothside_pass_files`
+Array of variant lists with bothside SR support for all batches, generated in [GenotypeBatch](./gb#sr_bothside_pass).
+
+#### `raw_sr_background_fail_files`
+Array of variant lists with low SR signal-to-noise ratio for all batches, generated in [GenotypeBatch](./gb#sr_background_fail).
+
+#### `localize_shard_size`
+Shard size for parallel computations. Decreasing this parameter may help reduce run time.
+
+#### `min_sr_background_fail_batches`
+Threshold fraction of batches with high SR background for a given variant required in order to assign this
+`HIGH_SR_BACKGROUND` flag. Most users should leave this at the default value.
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `combined_vcfs`
+Array of contig-sharded VCFs of combined PE/SR and depth calls.
+
+#### `cluster_bothside_pass_lists`
+Array of contig-sharded bothside SR support variant lists.
+
+#### `cluster_background_fail_lists`
+Array of contig-sharded high SR background variant lists.
+
+#### `combine_batches_merged_vcf`
+Genome-wide VCF of combined PE/SR and depth calls. Only generated if using [merge_vcfs](#optional--merge_vcfs).
diff --git a/website/docs/modules/concordance.md b/website/docs/modules/concordance.md
new file mode 100644
index 000000000..16e8814ec
--- /dev/null
+++ b/website/docs/modules/concordance.md
@@ -0,0 +1,58 @@
+---
+title: SVConcordance
+description: Annotates concordance with raw calls
+sidebar_position: 18
+slug: svc
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/SVConcordance.wdl)
+
+Annotates variants with genotype concordance against another SV call set. This is a general-purpose workflow that can
+be applied to any pair of VCFs containing the same sample set. This is also a prerequisite step for genotype filtering
+in the recommended pipeline: genotypes are compared to calls emitted by raw callers, where low concordance can be indicative
+of poor quality variants. See
+[GATK-SVConcordance](https://gatk.broadinstitute.org/hc/en-us/articles/27007917991707-SVConcordance-BETA) for more
+information on methods.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ amvf: ApplyManualVariantFilter
+ jrc: JoinRawCalls
+ svc: SVConcordance
+ fg: FilterGenotypes
+ amvf --> svc
+ jrc --> svc
+ svc --> fg
+
+ class svc thisModule
+ class amvf inModules
+ class jrc inModules
+ class fg outModules
+```
+
+### Inputs
+
+#### `output_prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `eval_vcf`
+VCF to annotate. In the recommended pipeline, this is generated in [ApplyManualVariantFilter](./amvf).
+
+#### `truth_vcf`
+VCF to compare against. This should contain the same samples as `eval_vcf`. In the recommended pipeline, this is
+generated in [JoinRawCalls](./jrc).
+
+### Outputs
+
+#### `concordance_vcf`
+"Eval" VCF annotated with genotype concordance.
+
diff --git a/website/docs/modules/downstream_filter.md b/website/docs/modules/downstream_filter.md
deleted file mode 100644
index f06df455b..000000000
--- a/website/docs/modules/downstream_filter.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-title: Downstream Filtering
-description: Downstream filtering (work in progress)
-sidebar_position: 12
-slug: df
----
-
-Apply downstream filtering steps to the cleaned VCF to further
-control the false discovery rate; all steps are optional
-and users should decide based on the specific purpose of
-their projects.
-
-Filtering methods include:
-
-- minGQ - remove variants based on the genotype quality across
- populations. Note: Trio families are required to build the minGQ
- filtering model in this step. We provide tables pre-trained with
- the 1000 genomes samples at different FDR thresholds for projects
- that lack family structures, and they can be found at the paths below.
- These tables assume that GQ has a scale of [0,999], so they will
- not work with newer VCFs where GQ has a scale of [0,99].
-
- ```shell
- gs://gatk-sv-resources-public/hg38/v0/sv-resources/ref-panel/1KG/v2/mingq/1KGP_2504_and_698_with_GIAB.10perc_fdr.PCRMINUS.minGQ.filter_lookup_table.txt
- gs://gatk-sv-resources-public/hg38/v0/sv-resources/ref-panel/1KG/v2/mingq/1KGP_2504_and_698_with_GIAB.1perc_fdr.PCRMINUS.minGQ.filter_lookup_table.txt
- gs://gatk-sv-resources-public/hg38/v0/sv-resources/ref-panel/1KG/v2/mingq/1KGP_2504_and_698_with_GIAB.5perc_fdr.PCRMINUS.minGQ.filter_lookup_table.txt
- ```
-
-- BatchEffect - remove variants that show significant discrepancies
- in allele frequencies across batches
-
-- FilterOutlierSamplesPostMinGQ - remove outlier samples with unusually
- high or low number of SVs
-
-- FilterCleanupQualRecalibration - sanitize filter columns and
- recalibrate variant QUAL scores for easier interpretation
diff --git a/website/docs/modules/evidence_qc.md b/website/docs/modules/evidence_qc.md
index 5177636da..a48b3e0a8 100644
--- a/website/docs/modules/evidence_qc.md
+++ b/website/docs/modules/evidence_qc.md
@@ -7,31 +7,28 @@ slug: eqc
import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/EvidenceQC.wdl)
+
Runs ploidy estimation, dosage scoring, and optionally VCF QC.
The results from this module can be used for QC and batching.
For large cohorts, this workflow can be run on arbitrary cohort
-partitions of up to about 500 samples. Afterwards, we recommend
+partitions of up to about 500 samples. Afterward, we recommend
using the results to divide samples into smaller batches (~100-500 samples)
-with ~1:1 male:female ratio. Refer to the [Batching](/docs/run/joint#batching) section
+with ~1:1 male:female ratio. Refer to the [Batching](/docs/execution/joint#batching) section
for further guidance on creating batches.
We also recommend using sex assignments generated from the ploidy
estimates and incorporating them into the PED file, with sex = 0 for sex aneuploidies.
-The following diagram illustrates the upstream and downstream workflows of the `EvidenceQC` workflow
-in the recommended invocation order. You may refer to
-[this diagram](https://github.com/broadinstitute/gatk-sv/blob/main/terra_pipeline_diagram.jpg)
-for the overall recommended invocation order.
-
-
+The following diagram illustrates the recommended invocation order:
```mermaid
stateDiagram
direction LR
- classDef inModules stroke-width:0px,fill:#00509d,color:#caf0f8
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
@@ -47,50 +44,83 @@ stateDiagram
class batching outModules
```
-
+### Preliminary Sample QC
+
+The purpose of sample filtering at this stage after EvidenceQC is to
+prevent very poor quality samples from interfering with the results for
+the rest of the callset. In general, samples that are borderline are
+okay to leave in, but you should choose filtering thresholds to suit
+the needs of your cohort and study. There will be future opportunities
+(as part of [FilterBatch](/docs/modules/fb)) for filtering before the joint genotyping
+stage if necessary. Here are a few of the basic QC checks that we recommend:
+
+- Chromosome X and Y ploidy plots: check that sex assignments
+ match your expectations. If there are discrepancies, check for
+ sample swaps and update your PED file before proceeding.
+
+- Whole-genome dosage score (WGD): examine distribution and check that
+ it is centered around 0 (the distribution of WGD for PCR-
+ samples is expected to be slightly lower than 0, and the distribution
+ of WGD for PCR+ samples is expected to be slightly greater than 0.
+ Refer to the gnomAD-SV paper for more information on WGD score).
+ Optionally filter outliers.
+
+- Low outliers for each SV caller: these are samples with
+ much lower than typical numbers of SV calls per contig for
+ each caller. An empty low outlier file means there were
+ no outliers below the median and no filtering is necessary.
+ Check that no samples had zero calls.
+
+- High outliers for each SV caller: optionally
+ filter outliers; samples with many more SV calls than average may be poor quality.
+
+- Remove samples with autosomal aneuploidies based on
+ the per-batch binned coverage plots of each chromosome.
### Inputs
-- Read count files (GatherSampleEvidence)
-- (Optional) SV call VCFs (GatherSampleEvidence)
+All array inputs of sample data must match in order. For example, the order of the `samples` array should match that
+of the `counts` array.
-### Outputs
+#### `batch`
+A name for the batch of samples being run. Can be alphanumeric with underscores.
-- Per-sample dosage scores with plots
-- Median coverage per sample
-- Ploidy estimates, sex assignments, with plots
-- (Optional) Outlier samples detected by call counts
+#### `samples`
+Sample IDs. Must match those used in [GatherSampleEvidence](./gse#outputs).
-## Preliminary Sample QC
+#### `counts`
+Binned read counts (`.counts.tsv.gz`) from [GatherSampleEvidence](./gse#outputs)
-The purpose of sample filtering at this stage after EvidenceQC is to
-prevent very poor quality samples from interfering with the results for
-the rest of the callset. In general, samples that are borderline are
-okay to leave in, but you should choose filtering thresholds to suit
-the needs of your cohort and study. There will be future opportunities
-(as part of FilterBatch) for filtering before the joint genotyping
-stage if necessary. Here are a few of the basic QC checks that we recommend:
+#### `*_vcfs`
+Raw SV call VCFs (`.vcf.gz`) from [GatherSampleEvidence](./gse#outputs). May be omitted in case a caller was not run.
-- Look at the X and Y ploidy plots, and check that sex assignments
- match your expectations. If there are discrepancies, check for
- sample swaps and update your PED file before proceeding.
+#### Optional `run_vcf_qc`
+Default: `false`. Run raw call VCF QC analysis.
-- Look at the dosage score (WGD) distribution and check that
- it is centered around 0 (the distribution of WGD for PCR-
- samples is expected to be slightly lower than 0, and the distribution
- of WGD for PCR+ samples is expected to be slightly greater than 0.
- Refer to the gnomAD-SV paper for more information on WGD score).
- Optionally filter outliers.
+#### Optional `run_ploidy`
+Default: `true`. Run ploidy estimation.
-- Look at the low outliers for each SV caller (samples with
- much lower than typical numbers of SV calls per contig for
- each caller). An empty low outlier file means there were
- no outliers below the median and no filtering is necessary.
- Check that no samples had zero calls.
+#### Optional `melt_insert_size`
+Mean insert size for each sample. Produces QC tables and plots if available.
-- Look at the high outliers for each SV caller and optionally
- filter outliers; samples with many more SV calls than average may be poor quality.
-- Remove samples with autosomal aneuploidies based on
- the per-batch binned coverage plots of each chromosome.
+### Outputs
+
+#### `WGD_*`
+Per-sample whole-genome dosage scores with plots
+
+#### `bincov_median`
+Median coverage per sample
+
+#### `bincov_matrix`
+Binned read depth matrix for the submitted batch
+
+#### `ploidy_*`
+Ploidy estimates, sex assignments, with plots
+
+#### Optional `*_qc_low`, `*_qc_high`
+Outlier samples detected by call counts.
+
+#### Optional `qc_table`
+QC summary table. Enable with [run_ploidy](#optional-run_ploidy).
diff --git a/website/docs/modules/filter_batch.md b/website/docs/modules/filter_batch.md
index 307c1acdd..9645a6223 100644
--- a/website/docs/modules/filter_batch.md
+++ b/website/docs/modules/filter_batch.md
@@ -5,36 +5,99 @@ sidebar_position: 7
slug: fb
---
-Filters poor quality variants and filters outlier samples.
-This workflow can be run all at once with the WDL at wdl/FilterBatch.wdl,
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/FilterBatch.wdl)
+
+Filters poor quality variants and outlier samples.
+This workflow can be run all at once with the top-level WDL,
or it can be run in three steps to enable tuning of outlier
filtration cutoffs. The three subworkflows are:
-1. FilterBatchSites: Per-batch variant filtration
+1. [FilterBatchSites](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/FilterBatchSites.wdl): Per-batch variant filtration
-2. PlotSVCountsPerSample: Visualize SV counts per
+2. [PlotSVCountsPerSample](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/PlotSVCountsPerSample.wdl): Visualize SV counts per
sample per type to help choose an IQR cutoff for
outlier filtering, and preview outlier samples for a given cutoff
-3. FilterBatchSamples: Per-batch outlier sample filtration;
- provide an appropriate outlier_cutoff_nIQR based on the
+3. [FilterBatchSamples](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/FilterBatchSamples.wdl): Per-batch outlier sample filtration;
+ provide an appropriate [outlier_cutoff_nIQR](#outlier_cutoff_niqr) based on the
SV count plots and outlier previews from step 2. Note
that not removing high outliers can result in increased
compute cost and a higher false positive rate in later steps.
-### Prerequisites
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
-- Generate Batch Metrics
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ gbm: GenerateBatchMetrics
+ fb: FilterBatch
+ mbs: MergeBatchSites
+ gbm --> fb
+ fb --> mbs
+
+ class fb thisModule
+ class gbm inModules
+ class mbs outModules
+```
### Inputs
-- Batch PED file
-- Metrics file (GenerateBatchMetrics)
-- Clustered SV and depth-only call VCFs (ClusterBatch)
+#### `batch`
+An identifier for the batch. Should match the name used in [GatherBatchEvidence](./gbe#batch).
+
+#### `*_vcf`
+Clustered VCFs from [ClusterBatch](./cb#clustered__vcf)
+
+#### `evidence_metrics`
+Metrics table [GenerateBatchMetrics](./gbm#metrics)
+
+#### `evidence_metrics_common`
+Common variant metrics table [GenerateBatchMetrics](./gbm#metrics_common)
+
+#### `outlier_cutoff_nIQR`
+Defines outlier sample cutoffs based on variant counts. Samples deviating from the batch median count by more than
+the given multiple of the interquartile range are hard filtered from the VCF. Recommended range is between `3` and `9`
+depending on desired sensitivity (higher is less stringent), or disable with `10000`.
+
+#### Optional `outlier_cutoff_table`
+A cutoff table to set permissible nIQR ranges for each SVTYPE. If provided, overrides `outlier_cutoff_nIQR`. Expected
+columns are: `algorithm`, `svtype`, `lower_cuff`, `higher_cff`. See the `outlier_cutoff_table` resource in
+[this json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/ref_panel_1kg.json) for an example table.
### Outputs
-- Filtered SV (non-depth-only a.k.a. "PESR") VCF with outlier samples excluded
-- Filtered depth-only call VCF with outlier samples excluded
-- Random forest cutoffs file
-- PED file with outlier samples excluded
\ No newline at end of file
+#### `filtered_depth_vcf`
+Depth-based CNV caller VCFs after variant and sample filtering.
+
+#### `filtered_pesr_vcf`
+PE/SR (non-depth) caller VCFs after variant and sample filtering.
+
+#### `cutoffs`
+Variant metric cutoffs for genotyping.
+
+#### `sv_counts`
+Array of TSVs containing SV counts for each sample, i.e. `sample-svtype-count` triplets. Each file corresponds to
+a different SV caller.
+
+#### `sv_count_plots`
+Array of images plotting SV counts stratified by SV type. Each file corresponds to a different SV caller.
+
+#### `outlier_samples_excluded`
+Array of sample IDs excluded by outlier analysis.
+
+#### `outlier_samples_excluded_file`
+Text file of sample IDs excluded by outlier analysis.
+
+#### `batch_samples_postOutlierExclusion`
+Array of remaining sample IDs after outlier exclusion.
+
+#### `batch_samples_postOutlierExclusion_file`
+Text file of remaining sample IDs after outlier exclusion.
diff --git a/website/docs/modules/filter_genotypes.md b/website/docs/modules/filter_genotypes.md
new file mode 100644
index 000000000..8f9ac445d
--- /dev/null
+++ b/website/docs/modules/filter_genotypes.md
@@ -0,0 +1,175 @@
+---
+title: FilterGenotypes
+description: Recalibrates qualities and filters genotypes
+sidebar_position: 19
+slug: fg
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/FilterGenotypes.wdl)
+
+Performs genotype quality recalibration using a machine learning model based on [xgboost](https://github.com/dmlc/xgboost)
+and filters genotypes. The output VCF contains the following updated fields:
+
+- `SL` : Scaled logit scores (see [here](#sl-scores))
+- `GQ` : Updated genotype quality rescaled using `SL`
+- `OGQ` : Original `GQ` score before recalibration
+- `HIGH_NCR` : Filter status assigned to variants exceeding a [threshold proportion](#optional-no_call_rate_cutoff)
+of no-call genotypes. This will also be applied to variants with genotypes that have already been filtered in the input VCF.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ svc: SVConcordance
+ fg: FilterGenotypes
+ avcf: AnnotateVcf
+ svc --> fg
+ fg --> avcf
+
+ class fg thisModule
+ class svc inModules
+ class avcf outModules
+```
+
+### Model features
+
+The model uses the following features:
+
+* Genotype properties:
+ * Non-reference and no-call allele counts
+ * Genotype quality (`GQ`)
+ * Supporting evidence types (`EV`) and respective genotype qualities (`PE_GQ`, `SR_GQ`, `RD_GQ`)
+ * Raw call concordance (`CONC_ST`)
+* Variant properties:
+ * Variant type (`SVTYPE`) and size (`SVLEN`)
+ * Calling algorithms (`ALGORITHMS`)
+ * Supporting evidence types (`EVIDENCE`)
+ * Two-sided SR support flag (`BOTHSIDES_SUPPORT`)
+ * Evidence overdispersion flag (`PESR_GT_OVERDISPERSION`)
+ * SR noise flag (`HIGH_SR_BACKGROUND`)
+ * Raw call concordance (`STATUS`, `NON_REF_GENOTYPE_CONCORDANCE`, `VAR_PPV`, `VAR_SENSITIVITY`, `TRUTH_AF`)
+* Reference context with respect to UCSC Genome Browser tracks:
+ * RepeatMasker
+ * Segmental duplications
+ * Simple repeats
+ * K-mer mappability (umap_s100 and umap_s24)
+
+### Model availability
+
+For ease of use, we provide a model pre-trained on high-quality data with truth data derived from long-read calls:
+```
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gatk-sv-recalibrator.aou_phase_1.v1.model
+```
+See the SV "Genotype Filter" section on page 34 of the [All of Us Genomic Quality Report C2022Q4R9 CDR v7](https://support.researchallofus.org/hc/en-us/articles/4617899955092-All-of-Us-Genomic-Quality-Report-ARCHIVED-C2022Q4R9-CDR-v7) for further details on model training. The generation and release of this model was made possible by the All of Us program (see [here](/docs/acknowledgements)).
+
+### SL scores
+
+All valid genotypes are annotated with a "scaled logit" (`SL`) score, which is rescaled to non-negative adjusted `GQ` values on [1, 99]. Note that the rescaled `GQ` values should *not* be interpreted as probabilities. Original genotype qualities are retained in the `OGQ` field.
+
+A more positive `SL` score indicates higher probability that the given genotype is not homozygous for the reference allele. Genotypes are therefore filtered using `SL` thresholds that depend on SV type and size. This workflow also generates QC plots using the [MainVcfQc](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MainVcfQc.wdl) workflow to review call set quality (see below for recommended practices).
+
+### Modes
+
+This workflow can be run in one of two modes:
+
+1. (Recommended) The user explicitly provides a set of `SL` cutoffs through the `sl_filter_args` parameter, e.g.
+ ```
+ "--small-del-threshold 93 --medium-del-threshold 150 --small-dup-threshold -51 --medium-dup-threshold -4 --ins-threshold -13 --inv-threshold -19"
+ ```
+ Genotypes with `SL` scores less than the cutoffs are set to no-call (`./.`). The above values were taken directly from Appendix N of the [All of Us Genomic Quality Report C2022Q4R9 CDR v7 ](https://support.researchallofus.org/hc/en-us/articles/4617899955092-All-of-Us-Genomic-Quality-Report-ARCHIVED-C2022Q4R9-CDR-v7). Users should adjust the thresholds depending on data quality and desired accuracy. Please see the arguments in [this script](https://github.com/broadinstitute/gatk-sv/blob/main/src/sv-pipeline/scripts/apply_sl_filter.py) for all available options.
+
+2. (Advanced) The user provides truth labels for a subset of non-reference calls, and `SL` cutoffs are automatically optimized. These truth labels should be provided as a json file in the following format:
+ ```json
+ {
+ "sample_1":
+ {
+ "good_variant_ids": ["variant_1", "variant_3"],
+ "bad_variant_ids": ["variant_5", "variant_10"]
+ },
+ "sample_2":
+ {
+ "good_variant_ids": ["variant_2", "variant_13"],
+ "bad_variant_ids": ["variant_8", "variant_11"]
+ }
+ }
+ ```
+ where "good_variant_ids" and "bad_variant_ids" are lists of variant IDs corresponding to non-reference (i.e. het or hom-var) sample genotypes that are true positives and false positives, respectively. `SL` cutoffs are optimized by maximizing the [F-score](https://en.wikipedia.org/wiki/F-score) with "beta" parameter `fmax_beta`, which modulates the weight given to precision over recall (lower values give higher precision).
+
+In both modes, the workflow additionally filters variants based on the "no-call rate", the proportion of genotypes that were filtered in a given variant. Variants exceeding the `no_call_rate_cutoff` are assigned a `HIGH_NCR` filter status.
+
+### QC recommendations
+
+We strongly recommend performing call set QC after this module. By default, QC plotting is enabled with the [run_qc](#optional-run_qc)
+argument. Users should carefully inspect the main plots from the [main_vcf_qc_tarball](#optional-main_vcf_qc_tarball).
+Please see the [MainVcfQc](./mvqc) module documentation for more information on interpreting these plots and recommended
+QC criteria.
+
+### Inputs
+
+#### Optional `vcf`
+Input VCF generated from [SVConcordance](./svc#concordance_vcf).
+
+#### Optional `output_prefix`
+Default: use input VCF filename. Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `ploidy_table`
+Table of sample ploidies generated in [JoinRawCalls](./jrc#ploidy_table).
+
+#### `gq_recalibrator_model_file`
+GQ-Recalibrator model. A public model is listed as `aou_recalibrate_gq_model_file` [here](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json).
+
+#### `recalibrate_gq_args`
+Arguments to pass to the `GQ` recalibration tool. Users should leave this with the default configuration in Terra.
+
+#### `genome_tracks`
+Genome tracks for sequence context annotation. Users should leave this with the default configuration in Terra.
+
+#### Optional `no_call_rate_cutoff`
+Default: `0.05`. Threshold fraction of samples that must have no-call genotypes in order to filter a variant. Set to 1 to disable.
+
+#### Optional `fmax_beta`
+Default: `0.4`. If providing a truth set, defines the beta parameter for F-score optimization.
+
+#### Optional `truth_json`
+Truth labels for input variants. If provided, the workflow will attempt to optimize filtering cutoffs automatically
+using the F-score. If provided, [sl_filter_args](#optional-sl_filter_args) is ignored.
+
+#### Optional `sl_filter_args`
+Arguments for the [SL filtering script](https://github.com/broadinstitute/gatk-sv/blob/main/src/sv-pipeline/scripts/apply_sl_filter.py).
+This should be used to set `SL` cutoffs for filtering (refer to description above). Overridden by [truth_json](#optional-truth_json).
+
+#### Optional `run_qc`
+Default: `true`. Enable running [MainVcfQc](./mvqc) automatically. By default, filtered variants will be excluded from
+the plots.
+
+#### Optional `optimize_vcf_records_per_shard`
+Default: `50000`. Shard size for scattered cutoff optimization tasks. Decrease this if those steps are running slowly.
+
+#### Optional `filter_vcf_records_per_shard`
+Default: `20000`. Shard size for scattered `GQ` recalibration tasks. Decrease this if those steps are running slowly.
+
+### Outputs
+
+#### `filtered_vcf`
+Filtered VCF.
+
+#### Optional `main_vcf_qc_tarball`
+QC plots generated with [MainVcfQc](./mvqc). Only generated if using [run_qc](#optional-run_qc).
+
+#### Optional `vcf_optimization_table`
+Table of cutoff optimization metrics. Only generated if [truth_json](#optional-truth_json) is provided.
+
+#### Optional `sl_cutoff_qc_tarball`
+Cutoff optimization and QC plots. Only generated if [truth_json](#optional-truth_json) is provided.
+
+#### `unfiltered_recalibrated_vcf`
+Supplemental output of the VCF after assigning `SL` genotype scores but before applying filtering.
\ No newline at end of file
diff --git a/website/docs/modules/gather_batch_evidence.md b/website/docs/modules/gather_batch_evidence.md
index 0bdf8a7a9..e06b75b8d 100644
--- a/website/docs/modules/gather_batch_evidence.md
+++ b/website/docs/modules/gather_batch_evidence.md
@@ -5,20 +5,21 @@ sidebar_position: 4
slug: gbe
---
-Runs CNV callers ([cn.MOPS](https://academic.oup.com/nar/article/40/9/e69/1136601), GATK-gCNV)
-and combines single-sample raw evidence into a batch.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
-The following diagram illustrates the downstream workflows of the `GatherBatchEvidence` workflow
-in the recommended invocation order. You may refer to
-[this diagram](https://github.com/broadinstitute/gatk-sv/blob/main/terra_pipeline_diagram.jpg)
-for the overall recommended invocation order.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/GatherBatchEvidence.wdl)
+
+Runs CNV callers ([cn.MOPS](https://academic.oup.com/nar/article/40/9/e69/1136601), [GATK-gCNV](https://github.com/broadinstitute/gatk))
+and combines single-sample raw evidence into batched files.
+
+The following diagram illustrates the recommended invocation order:
```mermaid
stateDiagram
direction LR
- classDef inModules stroke-width:0px,fill:#00509d,color:#caf0f8
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
@@ -34,148 +35,86 @@ stateDiagram
```
## Inputs
-This workflow takes as input the read counts, BAF, PE, SD, SR, and per-caller VCF files
-produced in the GatherSampleEvidence workflow, and contig ploidy and gCNV models from
-the TrainGCNV workflow.
-The following is the list of the inputs the GatherBatchEvidence workflow takes.
+:::info
+All array inputs of sample data must match in order. For example, the order of the `samples` array should match that of
+`counts`, `PE_files`, etc.
+:::
#### `batch`
-An identifier for the batch.
-
+An identifier for the batch; may only be alphanumeric with underscores.
#### `samples`
-Sets the list of sample IDs.
+Sample IDs. Must match the sample IDs used in [GatherSampleEvidence](./gse#sample_id) unless `rename_samples` is enabled, in
+which case sample IDs will be overwritten. See [sample ID requirements](/docs/gs/inputs#sampleids) for specifications
+of allowable sample IDs.
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
#### `counts`
-Set to the [`GatherSampleEvidence.coverage_counts`](./gse#coverage-counts) output.
-
-
-#### Raw calls
-
-The following inputs set the per-caller raw SV calls, and should be set
-if the caller was run in the [`GatherSampleEvidence`](./gse) workflow.
-You may set each of the following inputs to the linked output from
-the GatherSampleEvidence workflow.
-
-
-- `manta_vcfs`: [`GatherSampleEvidence.manta_vcf`](./gse#manta-vcf);
-- `melt_vcfs`: [`GatherSampleEvidence.melt_vcf`](./gse#melt-vcf);
-- `scramble_vcfs`: [`GatherSampleEvidence.scramble_vcf`](./gse#scramble-vcf);
-- `wham_vcfs`: [`GatherSampleEvidence.wham_vcf`](./gse#wham-vcf).
+Binned read count files (`*.rd.txt.gz`) generated in [GatherSampleEvidence](./gse#coverage-counts).
#### `PE_files`
-Set to the [`GatherSampleEvidence.pesr_disc`](./gse#pesr-disc) output.
+Discordant pair evidence files (`*.pe.txt.gz`) generated in [GatherSampleEvidence](./gse#pesr-disc).
#### `SR_files`
-Set to the [`GatherSampleEvidence.pesr_split`](./gse#pesr-split)
-
+Split read evidence files (`*.sr.txt.gz`) generated in [GatherSampleEvidence](./gse#pesr-split).
#### `SD_files`
-Set to the [`GatherSampleEvidence.pesr_sd`](./gse#pesr-sd)
-
-
-#### `matrix_qc_distance`
-You may refer to [this file](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/templates/terra_workspaces/cohort_mode/workflow_configurations/GatherBatchEvidence.json.tmpl)
-for an example value.
-
-
-#### `min_svsize`
-Sets the minimum size of SVs to include.
-
-
-#### `ped_file`
-A pedigree file describing the familial relationshipts between the samples in the cohort.
-Please refer to [this section](./#ped_file) for details.
+Site depth files (`*.sd.txt.gz`) generated in [GatherSampleEvidence](./gse#pesr-sd).
+#### `*_vcfs`
+Raw caller VCFs generated in [GatherSampleEvidence](./gse#outputs). Callers may be omitted if they were not run.
#### `run_matrix_qc`
-Enables or disables running optional QC tasks.
-
-
-#### `gcnv_qs_cutoff`
-You may refer to [this file](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/templates/terra_workspaces/cohort_mode/workflow_configurations/GatherBatchEvidence.json.tmpl)
-for an example value.
-
-#### cn.MOPS files
-The workflow needs the following cn.MOPS files.
-
-- `cnmops_chrom_file` and `cnmops_allo_file`: FASTA index files (`.fai`) for respectively
- non-sex chromosomes (autosomes) and chromosomes X and Y (allosomes).
- The file format is explained [on this page](https://www.htslib.org/doc/faidx.html).
-
- You may use the following files for these fields:
+Enables running QC tasks.
- ```json
- "cnmops_chrom_file": "gs://gcp-public-data--broad-references/hg38/v0/sv-resources/resources/v1/autosome.fai"
- "cnmops_allo_file": "gs://gcp-public-data--broad-references/hg38/v0/sv-resources/resources/v1/allosome.fai"
- ```
-
-- `cnmops_exclude_list`:
- You may use [this file](https://github.com/broadinstitute/gatk-sv/blob/d66f760865a89f30dbce456a3f720dec8b70705c/inputs/values/resources_hg38.json#L10)
- for this field.
-
-#### GATK-gCNV inputs
-
-The following inputs are configured based on the outputs generated in the [`TrainGCNV`](./gcnv) workflow.
-
-- `contig_ploidy_model_tar`: [`TrainGCNV.cohort_contig_ploidy_model_tar`](./gcnv#contig-ploidy-model-tarball)
-- `gcnv_model_tars`: [`TrainGCNV.cohort_gcnv_model_tars`](./gcnv#model-tarballs)
+#### `contig_ploidy_model_tar`
+Contig ploidy model tarball generated in [TrainGCNV](./gcnv#cohort_contig_ploidy_model_tar).
+#### `gcnv_model_tars`
+CNV model tarball generated in [TrainGCNV](./gcnv#cohort_gcnv_model_tars).
-The workflow also enables setting a few optional arguments of gCNV.
-The arguments and their default values are provided
-[here](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/templates/terra_workspaces/cohort_mode/workflow_configurations/GatherBatchEvidence.json.tmpl)
-as the following, and each argument is documented on
-[this page](https://gatk.broadinstitute.org/hc/en-us/articles/360037593411-PostprocessGermlineCNVCalls)
-and
-[this page](https://gatk.broadinstitute.org/hc/en-us/articles/360047217671-GermlineCNVCaller).
+#### Optional `rename_samples`
+Default: `false`. Overwrite sample IDs with the [samples](#samples) input.
+#### Optional `run_ploidy`
+Default: `false`. Runs ploidy estimation. Note this calls the same method used in [EvidenceQc](./eqc).
-#### Docker images
-
-The workflow needs the following Docker images, the latest versions of which are in
-[this file](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers.json).
+## Outputs
- - `cnmops_docker`;
- - `condense_counts_docker`;
- - `linux_docker`;
- - `sv_base_docker`;
- - `sv_base_mini_docker`;
- - `sv_pipeline_docker`;
- - `sv_pipeline_qc_docker`;
- - `gcnv_gatk_docker`;
- - `gatk_docker`.
+#### `merged_BAF`
+Batch B-allele frequencies file (`.baf.txt.gz`) derived from site depth evidence.
-#### Static inputs
+#### `merged_SR`
+Batch split read evidence file (`.sr.txt.gz`).
-You may refer to [this reference file](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json)
-for values of the following inputs.
+#### `merged_PE`
+Batch paired-end evidence file (`.pe.txt.gz`).
- - `primary_contigs_fai`;
- - `cytoband`;
- - `ref_dict`;
- - `mei_bed`;
- - `genome_file`;
- - `sd_locs_vcf`.
+#### `merged_bincov`
+Batch binned read counts file (`.rd.txt.gz`).
+#### `merged_dels`, `merged_dups`
+Batch CNV calls (`.bed.gz`).
-#### Optional Inputs
-The following is the list of a few optional inputs of the
-workflow, with an example of possible values.
+#### `median_cov`
+Median coverage table.
-- `"allosomal_contigs": [["chrX", "chrY"]]`
-- `"ploidy_sample_psi_scale": 0.001`
+#### `std_*_vcf_tar`
+Tarballs containing per-sample raw caller VCFs in standardized formats. This will be ommitted for any callers not
+provided in the inputs.
+#### Optional `batch_ploidy_*`
+Ploidy analysis files. Enabled with [run_ploidy](#optional-run_ploidy).
+#### Optional `*_stats`, `Matrix_QC_plot`
+QC files. Enabled with [run_matrix_qc](#run_matrix_qc).
+#### Optional `manta_tloc`
+Supplemental evidence for translocation variants. These records are hard filtered from the main call set but may be of
+interest to users investigating reciprocal translocations and other complex events.
-## Outputs
-- Combined read count matrix, SR, PE, and BAF files
-- Standardized call VCFs
-- Depth-only (DEL/DUP) calls
-- Per-sample median coverage estimates
-- (Optional) Evidence QC plots
diff --git a/website/docs/modules/gather_sample_evidence.md b/website/docs/modules/gather_sample_evidence.md
index fcc951be3..ca24b6a67 100644
--- a/website/docs/modules/gather_sample_evidence.md
+++ b/website/docs/modules/gather_sample_evidence.md
@@ -5,22 +5,26 @@ sidebar_position: 1
slug: gse
---
-Runs raw evidence collection on each sample with the following SV callers:
-Manta, Wham, Scramble, and/or MELT. For guidance on pre-filtering prior to GatherSampleEvidence,
-refer to the Sample Exclusion section.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
-The following diagram illustrates the downstream workflows of the `GatherSampleEvidence` workflow
-in the recommended invocation order. You may refer to
-[this diagram](https://github.com/broadinstitute/gatk-sv/blob/main/terra_pipeline_diagram.jpg)
-for the overall recommended invocation order.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/GatherSampleEvidence.wdl)
+Runs raw evidence collection (PE/SR/RD/SD) on each sample and performs SV discovery with the following callers:
+Manta, Wham, and Scramble. For guidance on pre-filtering prior to GatherSampleEvidence, refer to the
+[Input data](/docs/gs/inputs) section.
+
+:::note
+MELT is no longer supported as a raw caller. Please see [SV/CNV callers](/docs/gs/sv_callers) for more information.
+:::
+
+The following diagram illustrates the recommended invocation order:
```mermaid
stateDiagram
direction LR
- classDef inModules stroke-width:0px,fill:#00509d,color:#caf0f8
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
@@ -32,51 +36,104 @@ stateDiagram
class eqc outModules
```
-
-## Inputs
+### Inputs
#### `bam_or_cram_file`
-A BAM or CRAM file aligned to hg38. Index file (.bai) must be provided if using BAM.
+An indexed BAM or CRAM file aligned to hg38. See [input data requirements](/docs/gs/inputs).
#### `sample_id`
-Refer to the [sample ID requirements](/docs/gs/inputs#sampleids) for specifications of allowable sample IDs.
-IDs that do not meet these requirements may cause errors.
+Identifier string for the sample. Refer to the [sample ID requirements](/docs/gs/inputs#sampleids)
+for specifications of allowable sample IDs. IDs that do not meet these requirements may lead to errors.
+
+#### Optional `is_dragen_3_7_8`
+Default: detect automtically from BAM/CRAM header. The header check can be skippped by setting this parameter when it
+is known whether the BAM/CRAM is aligned with Dragen v3.7.8. If this is true and Scramble is configured to run then
+soft-clipped reads at sites called by Scramble in the original alignments will be realigned with BWA for re-calling with
+Scramble.
+
+#### Optional `collect_coverage` {#collect-coverage}
+Default: `true`. Collect read depth.
+
+#### Optional `collect_pesr` {#collect-pesr}
+Default: `true`. Collect paired-end (PE) split-read (SR), and site depth (SD) evidence.
+
+#### Optional `manta_docker` {#manta-docker}
+Manta docker image. If provided, runs the Manta tool.
+
+#### Optional `melt_docker` {#melt-docker}
+MELT docker image. If provided, runs the MELT tool.
+
+#### Optional `scramble_docker` {#scramble-docker}
+Scramble docker image. If provided, runs the Scramble tool.
+
+#### Optional `wham_docker` {#wham-docker}
+Wham docker image. If provided, runs the Wham tool.
+
+#### Optional `reference_bwa_*`
+BWA-mem index files. Required only if running Scramble and the input reads are aligned to Dragen v3.7.8
+
+#### Optional `scramble_alignment_score_cutoff`
+Default: `60` for Dragen v3.7.8 and `90` otherwise. Minimum alignment score for consensus sequences again the MEI reference
+in the Scramble tool. The default value is set automatically depending on aligner. Can be overridden to tune
+sensitivity.
+
+#### Optional `scramble_percent_align_cutoff`
+Default: `70`. Minimum alignment percent for consensus sequences again the MEI reference in the Scramble tool. Can be
+overridden to tune sensitivity.
+
+#### Optional `scramble_min_clipped_reads_fraction`
+Default: `0.22`. Minimum number of soft-clipped reads required for site cluster identification in the Scramble tool,
+as a fraction of average read depth. Can be overridden to tune sensitivity.
+
+### Advanced parameters
+
+#### Optional `run_localize_reads`
+Default: `false`. Copy input alignment files to the execution bucket before localizing to subsequent tasks. This
+may be desirable when BAM/CRAM files are stored in a requester-pays bucket or in another region to avoid egress charges.
+
+:::warning
+Enabling `run_localize_reads` can incur high storage costs. If using, make sure to clean up execution directories after
+the workflow finishes running.
+:::
-#### `preprocessed_intervals`
-Picard interval list.
+#### Optional `run_module_metrics`
+Default: `true`. Calculate QC metrics for the sample. If true, `primary_contigs_fai` must also be provided, and
+optionally the `baseline_*_vcf` inputs to run comparisons.
-#### `sd_locs_vcf`
-(`sd`: site depth)
-A VCF file containing allele counts at common SNP loci of the genome, which is used for calculating BAF.
-For human genome, you may use [`dbSNP`](https://www.ncbi.nlm.nih.gov/snp/)
-that contains a complete list of common and clinical human single nucleotide variations,
-microsatellites, and small-scale insertions and deletions.
-You may find a link to the file in
-[this reference](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json).
+#### Optional `move_bam_or_cram_files`
+Default: `false`. Uses `mv` instead of `cp` when operating on local CRAM/BAM files in some tasks. This can result in
+some performance improvement.
+:::warning
+Do not use `move_bam_or_cram_files` if running with a local backend or shared filesystem, as it may cause loss of
+input data.
+:::
-## Outputs
+### Outputs
-- Binned read counts file
-- Split reads (SR) file
-- Discordant read pairs (PE) file
+#### Optional `manta_vcf` {#manta-vcf}
+VCF containing variants called by Manta. Enabled by providing [manta_docker](#manta-docker).
-#### `manta_vcf` {#manta-vcf}
-A VCF file containing variants called by Manta.
+#### Optional `melt_vcf` {#melt-vcf}
+VCF containing variants called by MELT. Enabled by providing [melt_docker](#melt-docker).
-#### `melt_vcf` {#melt-vcf}
-A VCF file containing variants called by MELT.
+#### Optional `scramble_vcf` {#scramble-vcf}
+VCF containing variants called by Scramble. Enabled by providing [scramble_docker](#scramble-docker).
-#### `scramble_vcf` {#scramble-vcf}
-A VCF file containing variants called by Scramble.
+#### Optional `wham_vcf` {#wham-vcf}
+VCF containing variants called by Wham. Enabled by providing [wham_docker](#wham-docker).
-#### `wham_vcf` {#wham-vcf}
-A VCF file containing variants called by Wham.
+#### Optional `coverage_counts` {#coverage-counts}
+Binned read counts collected by `GATK-CollectReadCounts` (`*.counts.tsv.gz`). Enabled with [collect_coverage](#collect-coverage).
-#### `coverage_counts` {#coverage-counts}
+#### Optional `pesr_disc` {#pesr-disc}
+Discordant read pairs collected by `GATK-CollectSVEvidence` (`*.pe.txt.gz`). Enabled with [collect_pesr](#collect-pesr).
-#### `pesr_disc` {#pesr-disc}
+#### Optional `pesr_split` {#pesr-split}
+Split read positions collected by `GATK-CollectSVEvidence` (`*.sr.txt.gz`). Enabled with [collect_pesr](#collect-pesr).
-#### `pesr_split` {#pesr-split}
+#### Optional `pesr_sd` {#pesr-sd}
+Site depth counts collected by `GATK-CollectSVEvidence` (`*.sd.txt.gz`). Enabled with [collect_pesr](#collect-pesr).
-#### `pesr_sd` {#pesr-sd}
\ No newline at end of file
+#### Optional `sample_metrics_files`
+Sample metrics for QC. Enabled with [run_module_metrics](#optional-run_module_metrics).
\ No newline at end of file
diff --git a/website/docs/modules/generate_batch_metrics.md b/website/docs/modules/generate_batch_metrics.md
index 3f202e39b..d4488959b 100644
--- a/website/docs/modules/generate_batch_metrics.md
+++ b/website/docs/modules/generate_batch_metrics.md
@@ -5,19 +5,69 @@ sidebar_position: 6
slug: gbm
---
-Generates variant metrics for filtering.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/GenerateBatchMetrics.wdl)
-### Prerequisites
+Analyzes variants for RD, BAF, PE, and SR evidence and creates a table of metrics containing raw and statistical
+metrics. These results are used to assess variant quality in `FilterBatch` and for SR-based breakpoint refinement.
-- Cluster batch
+Modified tests are applied to common variants (carrier frequency at least 50%) and results are emitted in a separate table.
-### Inputs
+The following diagram illustrates the recommended invocation order:
-- Combined read count matrix, SR, PE, and BAF files (GatherBatchEvidence)
-- Per-sample median coverage estimates (GatherBatchEvidence)
-- Clustered SV VCFs (ClusterBatch)
-- Clustered depth-only call VCF (ClusterBatch)
+```mermaid
-### Outputs
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
-- Metrics file
+ cb: ClusterBatch
+ gbm: GenerateBatchMetrics
+ fb: FilterBatch
+ cb --> gbm
+ gbm --> fb
+
+ class gbm thisModule
+ class cb inModules
+ class fb outModules
+```
+
+## Inputs
+
+#### `batch`
+An identifier for the batch. Should match the name used in [GatherBatchEvidence](./gbe#batch).
+
+#### `*_vcf`
+Clustered VCFs from [ClusterBatch](./cb#clustered__vcf).
+
+#### `baf_metrics`
+Merged BAF evidence file from [GatherBatchEvidence](./gbe#merged_baf).
+
+#### `discfile`
+Merged PE evidence file from [GatherBatchEvidence](./gbe#merged_pe).
+
+#### `coveragefile`
+Merged RD evidence file from [GatherBatchEvidence](./gbe#merged_bincov).
+
+#### `splitfile`
+Merged SR evidence file from [GatherBatchEvidence](./gbe#merged_sr).
+
+#### `medianfile`
+Merged median coverage table from [GatherBatchEvidence](./gbe#median_cov).
+
+#### `*_split_size`
+Variants per shard for each evidence testing subworkflow. Reduce defaults to increase parallelism if the workflow is
+too slow.
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+## Outputs
+
+#### `metrics`
+TSV of variant metrics (excluding common variants).
+
+#### `metrics_common`
+TSV of common variant metrics (>50% carrier frequency).
diff --git a/website/docs/modules/genotype_batch.md b/website/docs/modules/genotype_batch.md
index 062aa9098..0c64733c8 100644
--- a/website/docs/modules/genotype_batch.md
+++ b/website/docs/modules/genotype_batch.md
@@ -5,23 +5,111 @@ sidebar_position: 9
slug: gb
---
-Genotypes a batch of samples across unfiltered variants combined across all batches.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/GenotypeBatch.wdl)
-### Prerequisites
-- Filter batch
-- Merge Batch Sites
+Genotypes a batch of samples across all variants in the cohort. Note that while the preceding step
+[MergeBatchSites](./msites) is a "cohort-level" module, genotyping is performed on one batch of samples at a time.
+
+In brief, genotyping is performed by first training variant metric cutoffs on sites with clear evidence signatures,
+and then genotypes and genotype qualities are assigned based on parametric models tuned with these cutoffs. This is
+performed separately for PE/SR calls and depth-based calls.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ mbs: MergeBatchSites
+ gb: GenotypeBatch
+ rgc: RegenotypeCNVs
+ mbs --> gb
+ gb --> rgc
+
+ class gb thisModule
+ class mbs inModules
+ class rgc outModules
+```
### Inputs
-- Batch PESR and depth VCFs (FilterBatch)
-- Cohort PESR and depth VCFs (MergeBatchSites)
-- Batch read count, PE, and SR files (GatherBatchEvidence)
+:::info
+A number of inputs to this module are only used in single-sample mode and therefore omitted here. In addition, some
+inputs marked as optional in the WDL are required for joint calling.
+:::
+
+#### `batch`
+An identifier for the batch. Should match the name used in [GatherBatchEvidence](./gbe#batch).
+
+#### `batch_pesr_vcf`
+Batch PE/SR caller variants after filtering, generated in [FilterBatch](./fb#filtered_pesr_vcf).
+
+#### `batch_depth_vcf`
+Batch depth caller variants after filtering, generated in [FilterBatch](./fb#filtered_depth_vcf).
+
+#### `cohort_pesr_vcf`
+Merged PE/SR caller variants for the cohort, generated in [MergeBatchSites](./msites#cohort_pesr_vcf).
+
+#### `cohort_depth_vcf`
+Merged depth caller variants for the cohort, generated in [MergeBatchSites](./msites#cohort_depth_vcf).
+
+#### `n_per_split`
+Records per shard when scattering variants. Decrease to increase parallelism if the workflow is running slowly.
+
+#### `coveragefile`
+Merged RD evidence file from [GatherBatchEvidence](./gbe#merged_bincov).
+
+#### `medianfile`
+Merged median coverage table from [GatherBatchEvidence](./gbe#median_cov).
+
+#### `rf_cutoffs`
+Genotyping cutoffs trained with the random forest filtering model from [FilterBatch](./fb#cutoffs).
+
+#### `seed_cutoffs`
+See [here](/docs/resources#seed_cutoffs).
+
+#### `n_RD_genotype_bins`
+Number of depth genotyping bins. Most users should leave this at the default value.
+
+#### `discfile`
+Merged PE evidence file from [GatherBatchEvidence](./gbe#merged_pe).
+
+#### `reference_build`
+Reference build version. Only "hg38" is supported.
+
+#### `sr_median_hom_ins`
+Median normalized split read counts of homozygous insertions. Most users should leave this at the default value.
+
+#### `sr_hom_cutoff_multiplier`
+Cutoff multiplier for split read counts of homozygous insertions. Most users should leave this at the default value.
### Outputs
-- Filtered SV (non-depth-only a.k.a. "PESR") VCF with outlier samples excluded
-- Filtered depth-only call VCF with outlier samples excluded
-- PED file with outlier samples excluded
-- List of SR pass variants
-- List of SR fail variants
-- (Optional) Depth re-genotyping intervals list
+#### `sr_bothside_pass`
+List of variant IDs with split reads found on both sides of the breakpoint.
+
+#### `sr_background_fail`
+List of variant IDs exhibiting low signal-to-noise ratio for split read evidence.
+
+#### `trained_PE_metrics`
+PE evidence genotyping metrics file.
+
+#### `trained_SR_metrics`
+SR evidence genotyping metrics file.
+
+#### `trained_genotype_*_*_sepcutoff`
+Trained genotyping cutoffs for variants called by PESR or depth when supported by PESR or depth evidence (4 total).
+
+#### `genotyped_depth_vcf`
+Genotyped depth call VCF.
+
+#### `genotyped_pesr_vcf`
+Genotyped PE/SR call VCF.
+
+#### `regeno_coverage_medians`
+Coverage metrics for downstream CNV regenotyping.
diff --git a/website/docs/modules/genotype_complex.md b/website/docs/modules/genotype_complex.md
new file mode 100644
index 000000000..8b4c88929
--- /dev/null
+++ b/website/docs/modules/genotype_complex.md
@@ -0,0 +1,89 @@
+---
+title: GenotypeComplexVariants
+description: Complex SV genotyping
+sidebar_position: 13
+slug: gcv
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/GenotypeComplexVariants.wdl)
+
+Genotypes, filters, and classifies putative complex variants using depth evidence.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ rcv: ResolveComplexVariants
+ gcv: GenotypeComplexVariants
+ cvcf: CleanVcf
+ rcv --> gcv
+ gcv --> cvcf
+
+ class gcv thisModule
+ class rcv inModules
+ class cvcf outModules
+```
+
+### Inputs
+
+:::info
+Some inputs of batch data must match in order. Specifically, the order of the `batches` array should match that of
+`depth_vcfs`, `bincov_files`, `depth_gt_rd_sep_files`, and `median_coverage_files`.
+:::
+
+#### `cohort_name`
+Cohort name. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `batches`
+Array of batch identifiers. Should match the name used in [GatherBatchEvidence](./gbe#batch).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+#### `depth_vcfs`
+Array of re-genotyped depth caller variants for all batches, generated in [RegenotypeCNVs](./rgcnvs#regenotyped_depth_vcfs).
+Must match order of [batches](#batches).
+
+#### Optional `merge_vcfs`
+Default: `false`. If true, merge contig-sharded VCFs into one genome-wide VCF. This may be used for convenience but cannot be used with
+downstream workflows.
+
+#### Optional `localize_shard_size`
+Default: `50000`. Shard size for parallel computations. Decreasing this parameter may help reduce run time.
+
+#### `complex_resolve_vcfs`
+Array of contig-sharded VCFs containing putative complex variants, generated in [ResolveComplexVariants](./rcv#complex_resolve_vcfs).
+
+#### `bincov_files`
+Array of RD evidence files for all batches from [GatherBatchEvidence](./gbe#counts). Must match order of [batches](#batches).
+
+#### `depth_gt_rd_sep_files`
+Array of "depth_depth" genotype cutoff files (depth evidence for depth-based calls) generated in
+[GenotypeBatch](./gb#trained_genotype___sepcutoff). Order must match that of [batches](#batches).
+
+#### `median_coverage_files`
+Array of median coverage tables for all batches from [GatherBatchEvidence](./gbe#median_cov). Order must match that of [batches](#batches).
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `complex_genotype_vcfs`
+Array of contig-sharded VCFs containing fully resolved and genotyped complex variants.
+
+#### `complex_genotype_merged_vcf`
+Genome-wide output VCF. Only generated if using [merge_vcfs](#optional--merge_vcfs).
diff --git a/website/docs/modules/index.md b/website/docs/modules/index.md
index a5f4ad7c5..e378cbbc4 100644
--- a/website/docs/modules/index.md
+++ b/website/docs/modules/index.md
@@ -4,50 +4,21 @@ description: Overview of the constituting components
sidebar_position: 0
---
-The pipeline is written in [Workflow Description Language (WDL)](https://openwdl.org),
-consisting of multiple modules to be executed in the following order.
+GATK-SV consists of multiple modules that need to be executed in a specific order. For joint calling,
+each module must be executed individually in sequence. While the single-sample mode invokes many of these modules, it is
+implemented as a single runnable workflow.
-- **GatherSampleEvidence** SV evidence collection, including calls from a configurable set of
- algorithms (Manta, MELT, and Wham), read depth (RD), split read positions (SR),
- and discordant pair positions (PE).
+The following diagram illustrates the overall module ordering:
-- **EvidenceQC** Dosage bias scoring and ploidy estimation.
+ -- **GatherBatchEvidence** Copy number variant calling using
- `cn.MOPS` and `GATK gCNV`; B-allele frequency (BAF) generation;
- call and evidence aggregation.
+Each module is implemented in the [Workflow Description Language (WDL)](https://openwdl.org). The Terra workspaces come
+pre-configured with default values for all required parameters and set up to run the pipeline for most use cases.
-- **ClusterBatch** Variant clustering
+The following sections supplement the Terra workspaces with documentation for each WDL, including an overview of its
+function, key input parameters, and outputs. Not all parameters are documented here, as some WDLs contain dozens of
+inputs. Descriptions of some common inputs can be found in the [Resource files](/docs/resources) and
+[Runtime attributes](/docs/runtime_attr) sections. Users are encouraged to refer to the WDL source code for additional
+clarification.
-- **GenerateBatchMetrics** Variant filtering metric generation
-
-- **FilterBatch** Variant filtering; outlier exclusion
-
-- **GenotypeBatch** Genotyping
-
-- **MakeCohortVcf** Cross-batch integration; complex variant resolution and re-genotyping; vcf cleanup
-
-- **Module 07 (in development)** Downstream filtering, including minGQ, batch effect check,
- outlier samples removal and final recalibration;
-
-- **AnnotateVCF** Annotations, including functional annotation,
- allele frequency (AF) annotation and AF annotation with external population callsets;
-
-- **Module 09 (in development)** Visualization, including scripts that generates IGV screenshots and rd plots.
-
-- Additional modules to be added: de novo and mosaic scripts
-
-
-## Pipeline Parameters
-
-Several inputs are shared across different modules of the pipeline, which are explained in this section.
-
-#### `ped_file`
-
-A pedigree file describing the familial relationships between the samples in the cohort.
-The file needs to be in the
-[PED format](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format).
-Updated with [EvidenceQC](./eqc) sex assignments, including
-`sex = 0` for sex aneuploidies;
-genotypes on chrX and chrY for samples with `sex = 0` in the PED file will be set to
-`./.` and these samples will be excluded from sex-specific training steps.
+For details on running GATK-SV on Terra, refer to the [Execution](/docs/execution/joint#instructions) section.
diff --git a/website/docs/modules/join_raw_calls.md b/website/docs/modules/join_raw_calls.md
new file mode 100644
index 000000000..55b563a24
--- /dev/null
+++ b/website/docs/modules/join_raw_calls.md
@@ -0,0 +1,54 @@
+---
+title: JoinRawCalls
+description: Clusters unfiltered variants across batches
+sidebar_position: 17
+slug: jrc
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/JoinRawCalls.wdl)
+
+This module clusters raw unfiltered variants from [ClusterBatch](./cb) across all batches. Concordance between these
+genotypes and the joint call set usually can be indicative of variant quality and is used downstream for genotype
+filtering.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cb: ClusterBatch
+ jrc: JoinRawCalls
+ svc: SVConcordance
+ cb --> jrc
+ jrc --> svc
+
+ class jrc thisModule
+ class cb inModules
+ class svc outModules
+```
+
+### Inputs
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. The guidelines outlined in the
+[sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `clustered_*_vcfs`
+Array of clustered VCFs for all batches, generated in [ClusterBatch](./cb#clustered__vcf).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+### Outputs
+
+#### `joined_raw_calls_vcf`
+VCF containing all raw calls in the cohort.
+
+#### `ploidy_table`
+TSV of contig ploidies for all samples, assuming diploid autosome and sex assignments from the [ped_file](#ped_file).
diff --git a/website/docs/modules/main_vcf_qc.md b/website/docs/modules/main_vcf_qc.md
new file mode 100644
index 000000000..d362765b4
--- /dev/null
+++ b/website/docs/modules/main_vcf_qc.md
@@ -0,0 +1,228 @@
+---
+title: MainVcfQC
+description: VCF QC plotting
+sidebar_position: 21
+slug: mvqc
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MainVcfQC.wdl)
+
+Creates plots for call set analysis and quality control (QC). This module is run by default with the
+[FilterGenotypes](./fg) module. Note, however, that the module is a stand-alone workflow and can be used with any batch-
+or cohort-level (i.e. multi-sample) SV VCF, including outputs from [ClusterBatch](./cb) and onward.
+
+Users can optionally provide external benchmarking datasets with the
+[site_level_comparison_datasets](#optional-site_level_comparison_datasets) and
+[sample_level_comparison_datasets](#optional-sample_level_comparison_datasets) parameters.
+
+The following sections provide guidance on interpreting these plots for VCFs that have been run with the recommended
+pipeline through [FilterGenotypes](./fg).
+
+### Output structure
+
+The tarball output has the following directory structure:
+
+- `/plots/main_plots` : main QC plots, described in detail [below](#plots-description).
+- `/plots/supplementary_plots` : comprehensive set of QC plots with greater detail, some of which are used as panels
+ in `main_plots`.
+- `/data` : various datafiles from the QC process such as family structures, a list of the samples
+ used for plotting, variant statistics, and lists of overlapping variants against any external call sets provided.
+
+### Plots description
+
+This section summarizes the most important plots used for overall call set QC found in the `/plots/main_plots`
+directory.
+
+Example plots are provided from a high-quality dataset consisting of 161 samples from the 1000 Genomes Project aligned
+with DRAGEN v3.7.8. Note that QC metrics and distributions will vary among data sets, particularly as sample size
+increases, and these examples are intended to provide a simple baseline for acceptable call set quality. Please see
+the [Recommendations](#recommendations) section for prescribed quality criteria.
+
+:::note
+The following plots are of variants passing all filters (i.e. with the `FILTER` status set to `PASS`). This is the
+default behavior of the QC plots generated in [FilterGenotypes](./fg).
+
+When running `MainVcfQc` as a standalone workflow, users may set the
+[bcftools_preprocessing_options](#optional--bcftools_preprocessing_options) argument to limit plotted variants based on `FILTER` status.
+For example, to limit to `PASS` variants for a VCF generated from [FilterGenotypes](./fg) use:
+```
+"bcftools_preprocessing_options": "-i 'FILTER~\"PASS\""
+```
+:::
+
+#### SV per genome
+
+Shows per-genome SV count distributions broken down by SV type. The top row is a site-level analysis, and
+the bottom row provides counts of individual alleles. Distributions of genotype quality (`GQ`), SV size, and
+frequency are also shown.
+
+
+
+#### SV counts
+
+Summarizes total variant counts by type, frequency, and size.
+
+
+
+#### Genotype distributions
+
+Distributions of genotypes across the call set.
+
+The left panel plots carrier frequency against allele frequency and can be used to visualize genotyping bias. In this
+plot, the rolling mean should not lie on the lower or upper extremes (all het. or all hom.).
+
+The middle panel plots the Hardy-Weinberg (HW) distribution of the cohort and provides estimates for the proportions of
+variants in HW equilibrium (or nominally in HW equilibrium before a multiple testing correction). The right-hand
+plots stratify HW by SV size.
+
+:::note
+The horizontal line bisecting the vertical axis in the AF vs. carrier frequency plot was fixed in.
+[#625](https://github.com/broadinstitute/gatk-sv/pull/625)).
+:::
+
+
+
+#### Size distributions
+
+Contains plots of SV length stratified by class and frequency.
+
+
+
+#### Frequency distributions
+
+Contains plots of allele frequency stratified by class and size.
+
+
+
+#### Optional SV trio inheritance
+
+Inheritance analysis of sites and alleles, including *de novo* rate distributions by SV class, size, frequency, and
+genotype quality. This plot is only generated if the cohort contains trios defined in the [PED file](#optional-ped_file).
+
+
+
+#### Optional Call set benchmarking
+
+This plot will be generated for each external call set provided. The left-hand panels show distributions of overlapping
+variants in the input VCF by SV class, size, and frequency. Colors depict proportions of variants meeting each overlap
+criterion. The right-hand panels show distributions among variants overlapping between the input and external call sets
+by SV class, size, and frequency.
+
+:::note
+Note the horizontal line bisecting the vertical axis in the AF vs. AF plot has been fixed in
+[#625](https://github.com/broadinstitute/gatk-sv/pull/625)).
+:::
+
+
+
+
+### Recommendations
+
+We suggest users observe the following basic criteria to assess the overall quality of the final call set:
+
+* Number of PASS variants per genome (which excludes `BND`) between 7,000 and 11,000.
+* Relative overall proportions of SV types similar to those shown above.
+* Similar size and frequency distributions to those shown above.
+* At least 70% of variants in HW equilibrium. Note that this may be lower depending on how well
+ the underlying cohort population structure adheres to the assumptions of the HW model. Note that HW equilibrium
+ usually improves after genotype filtering.
+* Strong allele frequency correlation against an external benchmarking dataset such as gnomAD-v2.
+* Low allele *de novo* inheritance rate (if trios are present), typically below 10%.
+
+These are intended as general guidelines and not strict limits on call set quality.
+
+### Inputs
+
+#### `vcfs`
+Array of one or more input VCFs. Each VCF must be indexed. This input is structured as an `Array` type for convenience
+when VCFs are sharded by contig. All VCFs in the array are concatenated before processing.
+
+#### Optional `vcf_format_has_cn`
+Default: `true`. This must be set to `false` for VCFs from modules before [CleanVcf](./cvcf) (i.e. without `CN` fields).
+
+#### Optional `bcftools_preprocessing_options`
+Additional arguments to pass to [bcftools view](https://samtools.github.io/bcftools/bcftools.html) before running
+the VCF through QC tools.
+
+#### Optional `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+If provided and there are trios present then [SV trio plots](#optional-sv-trio-inheritance) will be generated.
+
+#### Optional `list_of_samples_to_include`
+If provided, the input VCF(s) will be subset to samples in the list.
+
+#### Optional `sample_renaming_tsv`
+File with mapping to rename per-sample benchmark sample IDs for compatibility with cohort (see
+[here](/docs/resources#hgsv_byrska_bishop_sample_renaming_tsv) for example).
+
+#### Optional `max_trios`
+Default: `1000`. Upper limit on the number of trios to use for inheritance analysis.
+
+#### `prefix`
+Output prefix, such as cohort name. May be alphanumeric with underscores.
+
+#### `sv_per_shard`
+Records per shard for parallel processing. This parameter may be reduced if the workflow is running slowly.
+
+#### `samples_per_shard`
+Samples per shard for per-sample QC processing. This parameter may be reduced if the workflow is running slowly. Only
+has an effect when [do_per_sample_qc](#optional-do_per_sample_qc) is enabled.
+
+#### Optional `do_per_sample_qc`
+Default: `true`. If enabled, performs per-sample SV count plots, family analysis, and site-level benchmarking.
+
+#### Optional `site_level_comparison_datasets`
+Array of two-element arrays, one per dataset, each a tuple containing `[prefix, bed_uri_path]`, where the latter is
+a directory containing multiple bed files containing the call set stratified by subpopulation. For example:
+
+```
+["gnomAD_v2_Collins", "gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins"]
+```
+
+where the bucket directory contains the following files:
+
+```
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.AFR.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.ALL.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.AMR.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.EAS.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.EUR.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.OTH.bed.gz
+```
+
+Users can examine the above files as an example of how to format custom benchmarking datasets.
+
+See the `*_site_level_benchmarking_dataset` entries in the [Resource files](/docs/resources) section for available
+benchmarking call sets.
+
+#### Optional `sample_level_comparison_datasets`
+Array of two-element arrays, one per dataset, each a tuple containing `[prefix, tarball_uri]`, where the latter is
+a tarball containing sample-level benchmarking data. For example:
+
+```
+[["HGSV_ByrskaBishop", "gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/HGSV_ByrskaBishop_GATKSV_perSample.tar.gz"]]
+```
+
+Users can examine the above file as an example of how to format custom benchmarking datasets.
+
+See the `*_sample_level_benchmarking_dataset` entries in the [Resource files](/docs/resources#benchmarking-datasets)
+section for available benchmarking call sets.
+
+#### Optional `random_seed`
+Default: `0`. Random seed for sample subsetting in external call set comparisons.
+
+#### Optional `max_gq`
+Default: `99`. Max value to define range for `GQ` plotting. For modules prior to `CleanVcf`, use `999`.
+
+#### Optional `downsample_qc_per_sample`
+Default: `1000`. Number of samples to subset to for per-sample QC.
+
+### Outputs
+
+#### `sv_vcf_qc_output`
+Tarball of QC plots and data tables.
+
+#### `vcf2bed_output`
+Bed file containing all input variants.
diff --git a/website/docs/modules/make_cohort_vcf.md b/website/docs/modules/make_cohort_vcf.md
deleted file mode 100644
index d16e22acf..000000000
--- a/website/docs/modules/make_cohort_vcf.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-title: MakeCohortVcf
-description: Make Cohort VCF
-sidebar_position: 11
-slug: cvcf
----
-
-Combines variants across multiple batches, resolves complex variants,
-re-genotypes, and performs final VCF clean-up.
-
-### Prerequisites
-
-- GenotypeBatch
-- (Optional) RegenotypeCNVs
-
-### Inputs
-
-- RD, PE and SR file URIs (GatherBatchEvidence)
-- Batch filtered PED file URIs (FilterBatch)
-- Genotyped PESR VCF URIs (GenotypeBatch)
-- Genotyped depth VCF URIs (GenotypeBatch or RegenotypeCNVs)
-- SR pass variant file URIs (GenotypeBatch)
-- SR fail variant file URIs (GenotypeBatch)
-- Genotyping cutoff file URIs (GenotypeBatch)
-- Batch IDs
-- Sample ID list URIs
-
-### Outputs
-
-- Finalized "cleaned" VCF and QC plots
\ No newline at end of file
diff --git a/website/docs/modules/merge_batch_sites.md b/website/docs/modules/merge_batch_sites.md
index 6bfe43789..c1f789c09 100644
--- a/website/docs/modules/merge_batch_sites.md
+++ b/website/docs/modules/merge_batch_sites.md
@@ -5,16 +5,66 @@ sidebar_position: 8
slug: msites
---
-Combines filtered variants across batches. The WDL can be found at: /wdl/MergeBatchSites.wdl.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MergeBatchSites.wdl)
-### Prerequisites
-- Filter Batch
+Merges variants across batches. Variants are merged only if the following attributes match exactly:
+
+- Contig
+- Start position
+- End position (`END` field)
+- SV type (`SVTYPE` field)
+- SV length (`SVLEN` field, if available)
+- Strandedness (`STRANDS` field, if available)
+- Second contig (`CHR2` field, if available)
+- Second end (`END2` field, if available)
+
+This is a "cohort-level" workflow, meaning that is aggregates data across all batches. This is in contrast to all previous
+modules, which are sample- or batch-level. Note that this workflow should still be run on cohorts consisting of
+a single batch.
+
+:::info
+Terra users must configure a "sample_set_set" in their data table before running this module. See the [Execution
+section on MergeBatchSites](/docs/execution/joint#09-mergebatchsites) for futher instructions.
+:::
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ fb: FilterBatch
+ mbs: MergeBatchSites
+ gb: GenotypeBatch
+ fb --> mbs
+ mbs --> gb
+
+ class mbs thisModule
+ class fb inModules
+ class gb outModules
+```
### Inputs
-- List of filtered PESR VCFs (FilterBatch)
-- List of filtered depth VCFs (FilterBatch)
+#### `cohort`
+An identifier for the cohort. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids)
+section apply here.
+
+#### `depth_vcfs`
+Array of filtered depth VCFs across batches generated in [FilterBatch](./fb#filtered_depth_vcf).
+
+#### `pesr_vcfs`
+Array of filtered depth VCFs across batches generated in [FilterBatch](./fb#filtered_pesr_vcf).
### Outputs
-- Combined cohort PESR and depth VCFs
+#### `cohort_pesr_vcf`
+Merged PE/SR caller VCF.
+
+#### `cohort_depth_vcf`
+Merged depth caller VCF.
diff --git a/website/docs/modules/refine_cpx.md b/website/docs/modules/refine_cpx.md
new file mode 100644
index 000000000..6ed5e0f41
--- /dev/null
+++ b/website/docs/modules/refine_cpx.md
@@ -0,0 +1,84 @@
+---
+title: RefineComplexVariants
+description: Refines and filters complex variants
+sidebar_position: 15
+slug: refcv
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/RefineComplexVariants.wdl)
+
+Refines complex SVs and translocations and filters based on discordant read pair and read depth evidence reassessment.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cvcf: CleanVcf
+ refcv: RefineComplexVariants
+ amvf: ApplyManualVariantFilter
+
+ cvcf --> refcv
+ refcv --> amvf
+
+ class refcv thisModule
+ class cvcf inModules
+ class amvf outModules
+```
+
+### Inputs
+
+:::info
+All array inputs of batch data must match in order. For example, the order of the `batch_name_list` array should match
+that of `batch_sample_lists`, `PE_metrics`, etc.
+:::
+
+#### `vcf`
+Input vcf, generated in [CleanVcf](./cvcf#cleaned_vcf).
+
+#### `prefix`
+Prefix for output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `batch_name_list`
+Array of batch names. These should be the same batch names used in [GatherBatchEvidence](./gbe#batch).
+
+#### `batch_sample_lists`
+Array of sample ID lists for all batches, generated in [FilterBatch](./fb#batch_samples_postoutlierexclusion). Order must match [batch_name_list](#batch_name_list).
+
+#### `PE_metrics`
+Array of PE metrics files for all batches, generated in [GatherBatchEvidence](./gbe#merged_pe). Order must match [batch_name_list](#batch_name_list).
+
+#### `Depth_DEL_beds`, `Depth_DUP_beds`
+Arrays of raw DEL and DUP depth calls for all batches, generated in [GatherBatchEvidence](./gbe#merged_dels-merged_dups). Order must match [batch_name_list](#batch_name_list).
+
+#### `n_per_split`
+Shard size for parallel computations. Decreasing this parameter may help reduce run time.
+
+#### Optional `min_pe_cpx`
+Default: `3`. Minimum PE read count for complex variants (CPX).
+
+#### Optional `min_pe_ctx`
+Default: `3`. Minimum PE read count for translocations (CTX).
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `cpx_refined_vcf`
+Output VCF.
+
+#### `cpx_evidences`
+Supplementary output table of complex variant evidence.
diff --git a/website/docs/modules/regenotype_cnvs.md b/website/docs/modules/regenotype_cnvs.md
index cb5356b7e..b05dc2afe 100644
--- a/website/docs/modules/regenotype_cnvs.md
+++ b/website/docs/modules/regenotype_cnvs.md
@@ -1,24 +1,79 @@
---
-title: ReGenotypeCNVs
+title: RegenotypeCNVs
description: Regenotype CNVs
sidebar_position: 10
slug: rgcnvs
---
-Re-genotypes probable mosaic variants across multiple batches.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/RegenotypeCNVs.wdl)
-### Prerequisites
-- Genotype batch
+Re-genotypes probable mosaic variants across multiple batches. This is a "cohort-level" workflow that operates on
+all batches.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ gb: GenotypeBatch
+ rgc: RegenotypeCNVs
+ cb: CombineBatches
+ gb --> rgc
+ rgc --> cb
+
+ class rgc thisModule
+ class gb inModules
+ class cb outModules
+```
### Inputs
-- Per-sample median coverage estimates (GatherBatchEvidence)
-- Pre-genotyping depth VCFs (FilterBatch)
-- Batch PED files (FilterBatch)
-- Cohort depth VCF (MergeBatchSites)
-- Genotyped depth VCFs (GenotypeBatch)
-- Genotyped depth RD cutoffs file (GenotypeBatch)
+:::info
+All array inputs of batch data must match in order. For example, the order of the `batches` array should match that of
+`depth_vcfs`, `batch_depth_vcfs`, etc.
+:::
+
+#### `depth_vcfs`
+Array of genotyped depth caller variants for all batches, generated in [GenotypeBatch](./gb#genotyped_depth_vcf).
+
+#### `cohort_depth_vcf`
+Merged depth caller variants for the cohort, generated in [MergeBatchSites](./msites#cohort_depth_vcf).
+
+#### `batch_depth_vcfs`
+Array of filtered depth caller variants for all batches, generated in [FilterBatch](./fb#filtered_depth_vcf). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `coveragefiles`
+Array of merged RD evidence files for all batches from [GatherBatchEvidence](./gbe#merged_bincov). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `medianfiles`
+Array of median coverage tables for all batches from [GatherBatchEvidence](./gbe#median_cov). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `RD_depth_sepcutoffs`
+Array of "depth_depth" genotype cutoff files (depth evidence for depth-based calls) generated in
+[GenotypeBatch](./gb#trained_genotype___sepcutoff). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `n_per_split`
+Records per shard when scattering variants. Decrease to increase parallelism if the workflow is running slowly.
+
+#### `n_RD_genotype_bins`
+Number of depth genotyping bins. Most users should leave this at the default value.
+
+#### `batches`
+Array of batch identifiers. Should match the name used in [GatherBatchEvidence](./gbe#batch). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `cohort`
+Cohort name. May be alphanumeric with underscores.
+
+#### `regeno_coverage_medians`
+Array of regenotyping metrics generated in [GenotypeBatch](./gb#regeno_coverage_medians).
### Outputs
-- Re-genotyped depth VCFs.
+#### `regenotyped_depth_vcfs`
+Array of batch depth VCFs after regenotyping.
diff --git a/website/docs/modules/resolve_complex.md b/website/docs/modules/resolve_complex.md
new file mode 100644
index 000000000..5121e1a12
--- /dev/null
+++ b/website/docs/modules/resolve_complex.md
@@ -0,0 +1,89 @@
+---
+title: ResolveComplexVariants
+description: Complex SV discovery
+sidebar_position: 12
+slug: rcv
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/ResolveComplexVariants.wdl)
+
+Identifies multi-breakpoint complex variants, which are annotated with the `CPX` value in the `SVTYPE` field. These
+variants are putative, as read depth evidence is not assessed at this stage.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cb: CombineBatches
+ rcv: ResolveComplexVariants
+ gcv: GenotypeComplexVariants
+ cb --> rcv
+ rcv --> gcv
+
+ class rcv thisModule
+ class cb inModules
+ class gcv outModules
+```
+
+### Inputs
+
+:::info
+Some inputs of batch data must match in order. Specifically, the order of the `disc_files` array should match that of
+`rf_cutoff_files`.
+:::
+
+#### `cohort_name`
+Cohort name. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### Optional `merge_vcfs`
+Default: `false`. If true, merge contig-sharded VCFs into one genome-wide VCF. This may be used for convenience but cannot be used with
+downstream workflows.
+
+#### `cluster_vcfs`
+Array of contig-sharded VCFs, generated in [CombineBatches](./cmb#combined_vcfs).
+
+#### `cluster_bothside_pass_lists`
+Array of variant lists with bothside SR support for all batches, generated in [CombineBatches](./cmb#cluster_bothside_pass_lists).
+
+#### `cluster_background_fail_lists`
+Array of variant lists with low SR signal-to-noise ratio for all batches, generated in [CombineBatches](./cmb#cluster_background_fail_lists).
+
+#### `disc_files`
+Array of PE evidence files for all batches from [GatherBatchEvidence](./gbe#merged_pe).
+
+#### `rf_cutoffs`
+Array of batch genotyping cutoff files trained with the random forest filtering model from [FilterBatch](./fb#cutoffs).
+Must match the order of [disc_files](#disc_files).
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `complex_resolve_vcfs`
+Array of contig-sharded VCFs containing putative complex variants.
+
+#### `complex_resolve_bothside_pass_list`
+Array of contig-sharded bothside SR support variant lists.
+
+#### `complex_resolve_background_fail_list`
+Array of contig-sharded high SR background variant lists.
+
+#### `breakpoint_overlap_dropped_record_vcfs`
+Variants dropped due to exact overlap with another's breakpoint.
+
+#### `complex_resolve_merged_vcf`
+Genome-wide output VCF. Only generated if using [merge_vcfs](#optional--merge_vcfs).
diff --git a/website/docs/modules/train_gcnv.md b/website/docs/modules/train_gcnv.md
index 45c7b3cac..abd45ba8e 100644
--- a/website/docs/modules/train_gcnv.md
+++ b/website/docs/modules/train_gcnv.md
@@ -7,40 +7,33 @@ slug: gcnv
import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/TrainGCNV.wdl)
+
[GATK-gCNV](https://www.nature.com/articles/s41588-023-01449-0)
is a method for detecting rare germline copy number variants (CNVs)
from short-read sequencing read-depth information.
-The [TrainGCNV](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/TrainGCNV.wdl)
-module trains a gCNV model for use in the [GatherBatchEvidence](./gbe) workflow.
-The upstream and downstream dependencies of the TrainGCNV module are illustrated in the following diagram.
-
+The `TrainGCNV` module trains a gCNV model for use in the [GatherBatchEvidence](./gbe) workflow.
The samples used for training should be homogeneous (concerning sequencing platform,
-coverage, library preparation, etc.) and similar
-to the samples on which the model will be applied in terms of sample type,
-library preparation protocol, sequencer, sequencing center, and etc.
-
+coverage, library preparation, etc.) and similar to the samples on which the model will be applied.
For small, relatively homogeneous cohorts, a single gCNV model is usually sufficient.
However, for larger cohorts, especially those with multiple data sources,
we recommend training a separate model for each batch or group of batches (see
-[batching section](/docs/run/joint#batching) for details).
+[batching section](/docs/execution/joint#batching) for details).
The model can be trained on all or a subset of the samples to which it will be applied.
A subset of 100 randomly selected samples from the batch is a reasonable
input size for training the model; when the `n_samples_subsample` input is provided,
the `TrainGCNV` workflow can automatically perform this random selection.
-The following diagram illustrates the upstream and downstream workflows of the `TrainGCNV` workflow
-in the recommended invocation order. You may refer to
-[this diagram](https://github.com/broadinstitute/gatk-sv/blob/main/terra_pipeline_diagram.jpg)
-for the overall recommended invocation order.
+The following diagram illustrates the recommended invocation order:
```mermaid
stateDiagram
direction LR
- classDef inModules stroke-width:0px,fill:#00509d,color:#caf0f8
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
@@ -58,49 +51,59 @@ stateDiagram
## Inputs
-This section provides a brief description on the _required_ inputs of the TrainGCNV workflow.
-For a description on the _optional_ inputs and their default values, you may refer to the
-[source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/TrainGCNV.wdl) of the TrainGCNV workflow.
-Additionally, the majority of the optional inputs of the workflow map to the optional arguments of the
-tool the workflow uses, `GATK GermlineCNVCaller`; hence, you may refer to the
+The majority of the optional inputs of the workflow map to the optional arguments of the
+tool the workflow uses, `GATK-GermlineCNVCaller`; hence, you may refer to the
[documentation](https://gatk.broadinstitute.org/hc/en-us/articles/360040097712-GermlineCNVCaller)
-of the tool for a description on these optional inputs.
+of the tool for a description on these optional inputs. We recommend that most users use the defaults.
+
+:::info
+All array inputs of sample data must match in order. For example, the order of the `samples` array should match that
+of the `count_files` array.
+:::
#### `samples`
-A list of sample IDs.
-The order of IDs in this list should match the order of files in `count_files`.
+Sample IDs
#### `count_files`
-A list of per-sample coverage counts generated in the [GatherSampleEvidence](./gse#outputs) workflow.
+Per-sample binned read counts (`*.rd.txt.gz`) generated in the [GatherSampleEvidence](./gse#outputs) workflow.
-#### `contig_ploidy_priors`
-A tabular file with ploidy prior probability per contig.
-You may find the link to this input from
-[this reference](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json)
-and a description to the file format
-[here](https://gatk.broadinstitute.org/hc/en-us/articles/360037224772-DetermineGermlineContigPloidy).
+#### Optional `n_samples_subsample`, `sample_ids_training_subset`
+Provide one of these inputs to subset the input batch. `n_samples_subsample` will randomly subset, while
+`sample_ids_training_subset` is for defining a predetermined subset. These options are provided for convenience in Terra.
+## Outputs
-#### `reference_fasta`
-`reference_fasta`, `reference_index`, `reference_dict` are respectively the
-reference genome sequence in the FASTA format, its index file, and a corresponding
-[dictionary file](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format).
-You may find links to these files from
-[this reference](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json).
+#### `cohort_contig_ploidy_model_tar`
+Contig ploidy model tarball.
+#### `cohort_gcnv_model_tars`
+CNV model tarballs scattered across genomic intervals.
-## Outputs
+#### `cohort_contig_ploidy_calls_tar`
+Contig ploidy calls for the submitted batch.
-#### Optional `annotated_intervals` {#annotated-intervals}
+#### `cohort_gcnv_calls_tars`
+CNV call tarballs scattered by sample and genomic region prior to segmentation.
+#### `cohort_genotyped_segments_vcfs`
+Single-sample VCFs of CNV calls for the submitted batch.
+
+#### `cohort_gcnv_tracking_tars`
+Convergence tracking logs.
+
+#### `cohort_genotyped_intervals_vcfs`
+Single-sample VCFs for the submitted batch containing per-interval genotypes prior to segmentation.
+
+#### `cohort_denoised_copy_ratios`
+TSV files containing denoised copy ratios in each sample.
+
+#### Optional `annotated_intervals`
The count files from [GatherSampleEvidence](./gse) with adjacent intervals combined into
locus-sorted `DepthEvidence` files using `GATK CondenseDepthEvidence` tool, which are
annotated with GC content, mappability, and segmental-duplication content using
-[`GATK AnnotateIntervals`](https://gatk.broadinstitute.org/hc/en-us/articles/360041416652-AnnotateIntervals)
-tool. This output is generated if the optional input `do_explicit_gc_correction` is set to `True`.
-
-#### Optional `filtered_intervals_cnv` {#filtered-intervals-cnv}
-
-#### Optional `cohort_contig_ploidy_model_tar` {#contig-ploidy-model-tarball}
+[`GATK-AnnotateIntervals`](https://gatk.broadinstitute.org/hc/en-us/articles/360041416652-AnnotateIntervals)
+tool. This output is generated if `do_explicit_gc_correction` is set to `True`. Disabled by default.
-#### Optional `cohort_gcnv_model_tars` {#model-tarballs}
+#### Optional `filtered_intervals_cnv`, `filtered_intervals_ploidy`
+Intervals of read count bins to be used for CNV and ploidy calling after filtering for problematic regions (e.g.
+high GC content). This output is generated if `filter_intervals` is set to `True`. Enabled by default.
diff --git a/website/docs/modules/visualization.md b/website/docs/modules/visualization.md
deleted file mode 100644
index c6c71bf65..000000000
--- a/website/docs/modules/visualization.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-title: Visualization
-description: Visualization (work in progress)
-sidebar_position: 14
-slug: viz
----
-
-Visualize SVs with IGV screenshots and read depth plots.
-
-Visualization methods include:
-
-- RD Visualization - generate RD plots across all samples, ideal for visualizing large CNVs.
-- IGV Visualization - generate IGV plots of each SV for individual sample, ideal for visualizing de novo small SVs.
-- Module09.visualize.wdl - generate RD plots and IGV plots, and combine them for easy review.
diff --git a/website/docs/modules/visualize_cnvs.md b/website/docs/modules/visualize_cnvs.md
new file mode 100644
index 000000000..c0e792117
--- /dev/null
+++ b/website/docs/modules/visualize_cnvs.md
@@ -0,0 +1,51 @@
+---
+title: VisualizeCnvs
+description: Visualize CNVs
+sidebar_position: 22
+slug: viz
+---
+
+Generates plots of read depth across all samples. This is useful for visualizing large deletions and duplications
+(at least 5kbp).
+
+This module is not a part of the core pipeline but may be used to investigate variants of interest.
+
+### Inputs
+
+:::info
+All array inputs of batch data must match in order. In particular, the ordering of `median_files` and `rd_files` must
+be the same.
+:::
+
+#### `vcf_or_bed`
+VCF or bed file containing variants to plot. All variants will be automatically subsetted to DEL and DUP types subject
+to the [min_size](#min_size) constraint. VCF files must end in `.vcf.gz` and bed files must end in either `.bed` or
+`.bed.gz`. Bed files must contain columns: `chrom,start,end,name,svtype,samples`.
+
+#### `prefix`
+Output prefix, such as cohort name. May be alphanumeric with underscores.
+
+#### `median_files`
+Array of median coverage files for all batches in the input variants, generated in [GatherBatchEvidence](./gbe#median_cov).
+
+#### `rd_files`
+Array of RD evidence files for all batches in the input variants, generated in [GatherBatchEvidence](./gbe#merged_bincov).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+#### `min_size`
+Minimum size in bases of variants to plot.
+
+#### `flags`
+Additional flags to pass to the [RdTest plotting script](https://github.com/broadinstitute/gatk-sv/blob/main/src/RdTest/RdTest.R).
+
+:::warning
+Due to a bug, the `flags` parameter must contain `-s 999999999` in order to properly plot variants over 1 Mb.
+:::
+
+### Outputs
+
+#### `rdtest_plots`
+Tarball containing output plots.
+
diff --git a/website/docs/references.md b/website/docs/references.md
new file mode 100644
index 000000000..c30d1da62
--- /dev/null
+++ b/website/docs/references.md
@@ -0,0 +1,30 @@
+---
+title: Citation and references
+description: Citation and references
+sidebar_position: 11
+---
+
+### How to cite
+
+To credit GATK-SV, please cite the following publication:
+
+- [Collins RL et al. A structural variation reference for medical and population genetics. Nature. 2020 May;581(7809):444-451.](https://doi.org/10.1038/s41586-020-2287-8)
+
+### Additional references
+
+The following is a selected list of studies that have used GATK-SV:
+
+- [Belyeu JR et al. De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2,396 families. Am J Hum Genet. 2021 Apr 1;108(4):597-607.](https://doi.org/10.1016/j.ajhg.2021.02.012)
+- [Billingsley KJ et al. Genome-Wide Analysis of Structural Variants in Parkinson Disease. Ann Neurol. 2023 May;93(5):1012-1022.](https://doi.org/10.1002/ana.26608)
+- [Byrska-Bishop M et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell. 2022 Sep 1;185(18):3426-3440.e19.](https://doi.org/10.1016/j.cell.2022.08.004)
+- [Diaz Perez KK et al. Rare variants found in clinical gene panels illuminate the genetic and allelic architecture of orofacial clefting. Genet Med. 2023 Oct;25(10):100918.](https://doi.org/10.1016/j.gim.2023.100918)
+- [Gillani R et al. Rare germline structural variants increase risk for pediatric solid tumors. bioRxiv [Preprint]. 2024 Apr 29:2024.04.27.591484.](https://doi.org/10.1101/2024.04.27.591484)
+- [Han L et al. Functional annotation of rare structural variation in the human brain. Nat Commun. 2020 Jun 12;11(1):2990.](https://doi.org/10.1038/s41467-020-16736-1)
+- [Jurgens JA et al. Expanding the genetics and phenotypes of ocular congenital cranial dysinnervation disorders. Genet Med. 2024 Jul 17:101216.](https://doi.org/10.1016/j.gim.2024.101216)
+- [Kaivola K et al. Genome-wide structural variant analysis identifies risk loci for non-Alzheimer's dementias. Cell Genom. 2023 May 4;3(6):100316.](https://doi.org/10.1016/j.xgen.2023.100316)
+- [Koenig Z et al. A harmonized public resource of deeply sequenced diverse human genomes. Genome Res. 2024 Jun 25;34(5):796-809.](https://doi.org/10.1101/gr.278378.123)
+- [Lee AS et al. A cell type-aware framework for nominating non-coding variants in Mendelian regulatory disorders. Nat Commun. 2024 Sep 27;15(1):8268.](https://doi.org/10.1038/s41467-024-52463-7)
+- [Lowther C et al. Systematic evaluation of genome sequencing for the diagnostic assessment of autism spectrum disorder and fetal structural anomalies. Am J Hum Genet. 2023 Sep 7;110(9):1454-1469.](https://doi.org/10.1016/j.ajhg.2023.07.010)
+- [Werling DM et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet. 2018 Apr 26;50(5):727-736.](https://doi.org/10.1038/s41588-018-0107-y)
+- [Wojcik MH et al. Genome Sequencing for Diagnosing Rare Diseases. N Engl J Med. 2024 Jun 6;390(21):1985-1997.](https://doi.org/10.1056/nejmoa2314761)
+- [Zhao B et al. A neurodevelopmental disorder caused by a novel de novo SVA insertion in exon 13 of the SRCAP gene. Eur J Hum Genet. 2022 Sep;30(9):1083-1087.](https://doi.org/10.1038/s41431-022-01137-3)
diff --git a/website/docs/resources.md b/website/docs/resources.md
new file mode 100644
index 000000000..c27a13d5a
--- /dev/null
+++ b/website/docs/resources.md
@@ -0,0 +1,174 @@
+---
+title: Resource files
+description: Resource files manifest
+sidebar_position: 7
+---
+
+This page contains descriptions of common resource files used in the pipeline. All required files are publicly available
+in Google Cloud Storage Buckets. URIs for these files are available in the [hg38 resources json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json).
+
+:::info
+Some resources contain sensitive data and are stored in a secure bucket for development purposes. These files are not
+required to run GATK-SV.
+:::
+
+### Reference resources
+
+#### allosome_file
+Reference [fasta index file](https://www.htslib.org/doc/faidx.html) containing only allosomal contigs.
+
+#### autosome_file
+Reference [fasta index file](https://www.htslib.org/doc/faidx.html) containing only autosomal contigs.
+
+#### bin_exclude
+[Block-compressed](https://www.htslib.org/doc/bgzip.html) [bed file](https://genome.ucsc.edu/FAQ/FAQformat.html#format1)
+of intervals to exclude in the call set.
+
+#### cnmops_exclude_list
+Plain text bed file of N-masked regions in the reference.
+
+#### contig_ploidy_priors
+Plain text TSV file of prior probabilities contig ploidies used by GATK-gCNV.
+
+#### cytobands
+Block-compressed bed file of cytoband intervals.
+
+#### sd_locs_vcf
+Plain text VCF of SNP sites at which to collect site depth (SD) evidence.
+
+#### depth_exclude_list
+Block-compressed bed file of intervals over which to exclude overlapping depth-only calls.
+
+#### empty_file
+Empty file; used to satisfy some workflow code paths.
+
+#### exclude_intervals_for_gcnv_filter_intervals
+Plain text bed file of intervals to exclude from GATK-gCNV.
+
+#### external_af_ref_bed
+Block-compressed bed file of SV sites for external allele frequency annotation.
+
+#### genome_file
+Tab-delimited table of primary contigs with contig name in the first column and length in the second column.
+
+#### manta_region_bed
+Block-compressed bed file of intervals to call with Manta.
+
+#### mei_bed
+Block-compressed bed file of mobile element insertions in the reference genome.
+
+#### melt_std_vcf_header
+Text file containing the VCF header for raw MELT calls.
+
+#### noncoding_bed
+Plain text bed file of non-coding elements in the reference genome.
+
+#### par_bed
+Plain text bed file of pseudoautosomal regions.
+
+#### pesr_exclude_list
+Block-compressed bed file of intervals for filtering calls. Variants generated with non-CNV tools (Manta, MELT,
+Scramble, Wham) that have either end in any of these intervals are hard-filtered.
+
+#### preprocessed_intervals
+Intervals for read count collection and CNV calling.
+
+#### primary_contigs_fai
+Reference [fasta index file](https://www.htslib.org/doc/faidx.html) only containing the primary contigs, i.e. `chr1`, ...,
+`chr22`, `chrX`, and `chrY`.
+
+#### primary_contigs_list
+Text file of primary contig names.
+
+#### contigs_header
+Plain text VCF header section of primary contig sequences.
+
+#### protein_coding_gtf
+Protein coding sequence definitions for functional annotation in [General Transfer Format](https://www.ensembl.org/info/website/upload/gff.html).
+
+#### reference_dict
+Reference FASTA dictionary file (`*.dict`). See [this article](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format) for more information.
+
+#### reference_fasta
+Reference FASTA file (`*.fasta`). See [this article](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format) for more information.
+
+#### reference_index
+Reference FASTA index file (`*.fasta.fai`). See [this article](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format) for more information.
+
+#### rmsk
+Block-compressed bed file of [RepeatMasker intervals](https://genome.ucsc.edu/cgi-bin/hgTrackUi?g=rmsk).
+
+#### segdups
+Block-compressed bed file of [segmental duplication intervals](https://genome.ucsc.edu/cgi-bin/hgTrackUi?g=genomicSuperDups).
+
+#### seed_cutoffs
+TSV of cutoff priors for genotyping.
+
+#### single_sample_qc_definitions
+TSV of recommended ranges for single-sample QC metrics.
+
+#### wgd_scoring_mask
+Plain text bed file of whole-genome dosage (WGD) score intervals over which to assess coverage bias.
+
+#### wham_include_list_bed_file
+Plain text bed file of intervals to call with Wham.
+
+#### aou_recalibrate_gq_model_file
+Genotype filtering model trained using data generated by All of Us.
+
+#### hgdp_recalibrate_gq_model_file
+Genotype filtering model trained using data generated from the Human Genome Diversity Project.
+
+#### recalibrate_gq_genome_tracks
+List of block-compressed bed files, each containing intervals from a separate genome track.
+
+### Benchmarking datasets
+
+#### ccdg_abel_site_level_benchmarking_dataset
+Benchmarking variant set from [Abel et al. 2020](https://doi.org/10.1038/s41586-020-2371-0).
+
+#### gnomad_v2_collins_sample_level_benchmarking_dataset
+Benchmarking genotypes from gnomAD-SV-v2, see [Collins et al. 2020](https://doi.org/10.1038/s41586-020-2287-8). **Not public data.**
+
+#### gnomad_v2_collins_site_level_benchmarking_dataset
+Benchmarking variant set from gnomAD-SV-v2, see [Collins et al. 2020](https://doi.org/10.1038/s41586-020-2287-8).
+
+#### hgsv_byrska_bishop_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Byrska-Bishop et al. 2022](https://doi.org/10.1016/j.cell.2022.08.004).
+
+#### hgsv_byrska_bishop_sample_renaming_tsv
+Sample renaming manifest for the Byrska-Bishop benchmarking genotypes.
+
+#### hgsv_byrska_bishop_site_level_benchmarking_dataset
+Benchmarking variant set from [Byrska-Bishop et al. 2022](https://doi.org/10.1016/j.cell.2022.08.004).
+
+#### hgsv_ebert_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Ebert et al. 2021](https://doi.org/10.1126/science.abf7117).
+
+#### ssc_belyeu_sample_level_benchmarking_dataset
+Benchmarking genotypes from the [Simons Simplex Collection](https://www.sfari.org/resource/simons-simplex-collection/), derived from [Belyeu et al. 2021](https://doi.org/10.1016/j.ajhg.2021.02.012). **Not public data.**
+
+#### ssc_belyeu_site_level_benchmarking_dataset
+Benchmarking variant set from the [Simons Simplex Collection](https://www.sfari.org/resource/simons-simplex-collection/), derived from [Belyeu et al. 2021](https://doi.org/10.1016/j.ajhg.2021.02.012). **Not public data.**
+
+#### ssc_sanders_sample_level_benchmarking_dataset
+Benchmarking genotypes from the [Simons Simplex Collection](https://www.sfari.org/resource/simons-simplex-collection/), derived from [Sanders et al. 2015](https://doi.org/10.1016/j.neuron.2015.09.016). **Not public data.**
+
+#### thousand_genomes_site_level_benchmarking_dataset
+Benchmarking variant set from the 1000 Genomes Project Phase 3 SV call set, see [Sudmant et al. 2015](https://doi.org/10.1038/nature15394).
+
+#### asc_site_level_benchmarking_dataset
+Benchmarking variant set from the Autism Spectrum Consortium. **Not public data.**
+
+#### hgsv_site_level_benchmarking_dataset
+Benchmarking variant set from [Werling et al. 2018](https://doi.org/10.1038%2Fs41588-018-0107-y). **Not public data.**
+
+#### collins_2017_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Collins et al. 2017](https://doi.org/10.1186/s13059-017-1158-6). **Not public data.**
+
+#### sanders_2015_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Sanders et al. 2015](https://doi.org/10.1016/j.neuron.2015.09.016). **Not public data.**
+
+#### werling_2018_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Werling et al. 2018](https://doi.org/10.1038%2Fs41588-018-0107-y). **Not public data.**
+
diff --git a/website/docs/run/joint.md b/website/docs/run/joint.md
deleted file mode 100644
index 2d73959a3..000000000
--- a/website/docs/run/joint.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-title: Joint-calling
-description: Run the pipeline on a cohort
-sidebar_position: 4
-slug: joint
----
-
-:::info
-This documentation page is incomplete, and we are actively working on improving it with comprehensive information.
-:::
-
-## Batching
-
-For larger cohorts, samples should be split up into batches of about 100-500
-samples with similar characteristics. We recommend batching based on overall
-coverage and dosage score (WGD), which can be generated in [EvidenceQC](/docs/modules/eqc).
-An example batching process is outlined below:
-
-1. Divide the cohort into PCR+ and PCR- samples
-2. Partition the samples by median coverage from [EvidenceQC](/docs/modules/eqc),
- grouping samples with similar median coverage together. The end goal is to
- divide the cohort into roughly equal-sized batches of about 100-500 samples;
- if your partitions based on coverage are larger or uneven, you can partition
- the cohort further in the next step to obtain the final batches.
-3. Optionally, divide the samples further by dosage score (WGD) from
- [EvidenceQC](/docs/modules/eqc), grouping samples with similar WGD score
- together, to obtain roughly equal-sized batches of about 100-500 samples
-4. Maintain a roughly equal sex balance within each batch, based on sex
- assignments from [EvidenceQC](/docs/modules/eqc)
diff --git a/website/docs/run/overview.md b/website/docs/run/overview.md
deleted file mode 100644
index f9a82bcd1..000000000
--- a/website/docs/run/overview.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-title: Overview
-description: Overview
-sidebar_position: 1
-slug: overview
----
-
-There are two factors to consider when deciding how to run GATK-SV.
-
-
-1. **Variant calling modes: single-sample and cohort-based calling.**
- GATK-SV offers two distinct pipeline configurations for detecting
- structural variations (SVs), each tailored for different research needs:
-
- - **Single-sample analysis:**
- This configuration is ideal for examining SVs in individual samples,
- focusing exclusively on data from that single sample. Running this mode is less complex,
- involving just one workflow per sample.
-
- - **Joint calling:**
- This configuration is designed for more extensive studies, such as those
- involving population genetics or disease association studies.
- It analyzes SVs across a cohort by collectively assessing data from all samples.
- However, this comes with increased complexity compared to the single-sample mode,
- requiring the execution of multiple workflows and involves data preparation steps
- (e.g., batching files from the cohort).
-
-
-2. **Which platform you would like to use for running GATK-SV?**
- You may run GATK-SV on the following platforms.
-
- - [Terra.bio](https://terra.bio): A user-friendly cloud-native platform for scalable data analysis.
- The primary focus of this documentation is on supporting the execution of GATK-SV within the Terra platform.
-
- - [Cromwell](https://github.com/broadinstitute/cromwell):
- You may run GATK-SV on a self-hosted and managed cromwell instance, which is ideal for
- power-users and developers. We provide guidelines for this option in the
- [_advanced guides_](/docs/advanced/development/cromwell) section.
-
-
-Your decision regarding the execution modes and platform should be guided by
-the objectives of your study, the size of your cohort, data access needs,
-and the trade-off between a straightforward interface (Terra)
-and more detailed customization options (self-managed Cromwell server).
-Please refer to the following documentation on running GATK-SV within the Terra platform.
-
-- [Single-sample on Terra](single.md);
-- [Joint calling on Terra](joint.md).
diff --git a/website/docs/run/single.md b/website/docs/run/single.md
deleted file mode 100644
index 3f444b9d7..000000000
--- a/website/docs/run/single.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-title: Single-sample
-description: Run the pipeline on a single sample
-sidebar_position: 3
-slug: single
----
-
-:::info
-This documentation page is incomplete, and we are actively working on improving it with comprehensive information.
-:::
-
-We have developed a
- [Terra workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
-for running GATK-SV on a single sample, which contains the related documentation.
diff --git a/website/docs/runtime_attr.md b/website/docs/runtime_attr.md
new file mode 100644
index 000000000..be3cb6de2
--- /dev/null
+++ b/website/docs/runtime_attr.md
@@ -0,0 +1,51 @@
+---
+title: Runtime attributes
+description: Runtime attributes
+sidebar_position: 6
+---
+
+GATK-SV is implemented as a set of WDLs designed to run on Google Cloud Platform. Computations are broken up into
+a set of tasks that are carried out in a particular order on cloud virtual machines (VMs), each of which
+possesses a limited set of resources. These include the following primary components:
+
+- CPU cores
+- Random access memory (RAM)
+- Disk storage
+
+GATK-SV attempts to request the appropriate amount of resources for each task and in some cases employs mathematical
+models to predict optimal requirements that minimize cost. However, if the actual computations performed require more
+than the requested resources on the VM, the task may fail.
+
+:::info
+Most tasks in GATK-SV are tuned for cohorts with ~150 samples. Most tasks should scale automatically, but running larger
+cohorts may lead to slowdowns or errors due to insufficient resource allocation.
+:::
+
+In addition to VM resources, Terra and Cromwell both support automatic retries of tasks in case of ephemeral errors,
+as well as requesting [preemptible VMs](https://cloud.google.com/compute/docs/instances/preemptible) at a significant discount.
+
+### Setting runtime attributes
+
+GATK-SV exposes optional parameters for manually tuning VM resource allocation, automatic retries, and the use of preemptible
+instances. These parameters are all a custom struct called `RuntimeAttr`, which is defined in
+[Structs.wdl](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/Structs.wdl) as:
+
+```
+struct RuntimeAttr {
+ Float? mem_gb
+ Int? cpu_cores
+ Int? disk_gb
+ Int? boot_disk_gb
+ Int? preemptible_tries
+ Int? max_retries
+}
+```
+
+Users encountering errors due to insufficient VM resources or wishing to adjust automatic and preemptible retries may
+modify the corresponding parameters. Users should inspect the WDL source to determine whether a given `RuntimeAttr`
+parameter corresponds to a specific task.
+
+### Further reading
+
+For more information on identifying errors and setting runtime parameters, please refer to the
+[Troubleshooting FAQ](/docs/troubleshooting/faq).
diff --git a/website/docs/troubleshooting/_category_.json b/website/docs/troubleshooting/_category_.json
index 9bcfbef40..09047afec 100644
--- a/website/docs/troubleshooting/_category_.json
+++ b/website/docs/troubleshooting/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Troubleshooting",
- "position": 5,
+ "position": 8,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/troubleshooting/faq.md b/website/docs/troubleshooting/faq.md
index 1cd431028..439cbf391 100644
--- a/website/docs/troubleshooting/faq.md
+++ b/website/docs/troubleshooting/faq.md
@@ -3,12 +3,17 @@ title: FAQ
slug: faq
---
+Please consult the following resources for additional troubleshooting guides:
+
+- [Troubleshooting GATK-SV (article)](https://gatk.broadinstitute.org/hc/en-us/articles/5334566940699-Troubleshooting-GATK-SV)
+- [Troubleshooting GATK-SV Error Messages on Terra (video)](https://www.youtube.com/watch?v=3UVV03H9p1w)
+
### VM runs out of memory or disk
-- Default pipeline settings are tuned for batches of 100 samples.
+- Default pipeline settings are tuned for batches of 150 samples.
Larger batches or cohorts may require additional VM resources.
Most runtime attributes can be modified through
- the RuntimeAttr inputs. These are formatted like this in the json:
+ the `RuntimeAttr` inputs. These can be formatted like this in an input json:
```json
"MyWorkflow.runtime_attr_override": {
@@ -18,7 +23,7 @@ slug: faq
```
Note that a subset of the struct attributes can be specified.
- See `wdl/Structs.wdl` for available attributes.
+ See [wdl/Structs.wdl](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/Structs.wdl) for available attributes.
### Calculated read length causes error in MELT workflow
diff --git a/website/package.json b/website/package.json
index fa32a8117..70fea28a2 100644
--- a/website/package.json
+++ b/website/package.json
@@ -14,9 +14,9 @@
"write-heading-ids": "docusaurus write-heading-ids"
},
"dependencies": {
- "@docusaurus/core": "3.3.2",
- "@docusaurus/preset-classic": "3.3.2",
- "@docusaurus/theme-mermaid": "3.3.2",
+ "@docusaurus/core": "^3.5.2",
+ "@docusaurus/preset-classic": "^3.5.2",
+ "@docusaurus/theme-mermaid": "^3.5.2",
"@mdx-js/react": "^3.0.0",
"clsx": "^2.0.0",
"prism-react-renderer": "^2.3.0",
@@ -24,8 +24,8 @@
"react-dom": "^18.0.0"
},
"devDependencies": {
- "@docusaurus/module-type-aliases": "3.3.2",
- "@docusaurus/types": "3.3.2"
+ "@docusaurus/module-type-aliases": "^3.5.2",
+ "@docusaurus/types": "^3.5.2"
},
"browserslist": {
"production": [
diff --git a/website/static/img/qc/VcfQcGenotypeDistributions.png b/website/static/img/qc/VcfQcGenotypeDistributions.png
new file mode 100644
index 000000000..c18ae574d
--- /dev/null
+++ b/website/static/img/qc/VcfQcGenotypeDistributions.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:23e7e565ed6550ea79332bef7dea04a1fff2ed28932b96d694d8de5d7e690352
+size 1115174
diff --git a/website/static/img/qc/VcfQcGnomADv2CollinsSVAllCallsetBenchmarking.png b/website/static/img/qc/VcfQcGnomADv2CollinsSVAllCallsetBenchmarking.png
new file mode 100644
index 000000000..d89e0d16a
--- /dev/null
+++ b/website/static/img/qc/VcfQcGnomADv2CollinsSVAllCallsetBenchmarking.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:9cc3dfca00798620a6b2b90bbd51fd0b741800b9b4c768556b4d7aac763a1c9d
+size 1064628
diff --git a/website/static/img/qc/VcfQcSizeDistributionsMerged.png b/website/static/img/qc/VcfQcSizeDistributionsMerged.png
new file mode 100644
index 000000000..4d468cdf3
--- /dev/null
+++ b/website/static/img/qc/VcfQcSizeDistributionsMerged.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:561a681de80ee93f8a1dfc47b0086828dde6c121223c4ebff6d47a73e88697e8
+size 151637
diff --git a/website/static/img/qc/VcfQcSvCountsMerged.png b/website/static/img/qc/VcfQcSvCountsMerged.png
new file mode 100644
index 000000000..7d2c5fd77
--- /dev/null
+++ b/website/static/img/qc/VcfQcSvCountsMerged.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:64a882fe1ca04ec7da830fb249c2799e5fbab052951a476facfe2eca01c5214b
+size 40665
diff --git a/website/static/img/qc/VcfQcSvPerGenome.png b/website/static/img/qc/VcfQcSvPerGenome.png
new file mode 100644
index 000000000..15d06f19c
--- /dev/null
+++ b/website/static/img/qc/VcfQcSvPerGenome.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:b04d0ac119d7738f5c084db546df86d6c55d030ca72eea10807f390af4b87fe5
+size 702941
diff --git a/website/static/img/qc/VcfQcSvTrioInheritance.png b/website/static/img/qc/VcfQcSvTrioInheritance.png
new file mode 100644
index 000000000..44cc1ad59
--- /dev/null
+++ b/website/static/img/qc/VcfQcSvTrioInheritance.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:068adff3b217398fe4f410cdc70ac128a56bce44ea811f58114c518c9547a692
+size 134280
diff --git a/website/static/img/qc/VcfQqFreqDistributionsMerged.png b/website/static/img/qc/VcfQqFreqDistributionsMerged.png
new file mode 100644
index 000000000..7661d2eb2
--- /dev/null
+++ b/website/static/img/qc/VcfQqFreqDistributionsMerged.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:6618befbf0a05ebd683783995d974e0bb5720f3f41c39268c1c18f3b091154eb
+size 97067
-- **GatherBatchEvidence** Copy number variant calling using
- `cn.MOPS` and `GATK gCNV`; B-allele frequency (BAF) generation;
- call and evidence aggregation.
+Each module is implemented in the [Workflow Description Language (WDL)](https://openwdl.org). The Terra workspaces come
+pre-configured with default values for all required parameters and set up to run the pipeline for most use cases.
-- **ClusterBatch** Variant clustering
+The following sections supplement the Terra workspaces with documentation for each WDL, including an overview of its
+function, key input parameters, and outputs. Not all parameters are documented here, as some WDLs contain dozens of
+inputs. Descriptions of some common inputs can be found in the [Resource files](/docs/resources) and
+[Runtime attributes](/docs/runtime_attr) sections. Users are encouraged to refer to the WDL source code for additional
+clarification.
-- **GenerateBatchMetrics** Variant filtering metric generation
-
-- **FilterBatch** Variant filtering; outlier exclusion
-
-- **GenotypeBatch** Genotyping
-
-- **MakeCohortVcf** Cross-batch integration; complex variant resolution and re-genotyping; vcf cleanup
-
-- **Module 07 (in development)** Downstream filtering, including minGQ, batch effect check,
- outlier samples removal and final recalibration;
-
-- **AnnotateVCF** Annotations, including functional annotation,
- allele frequency (AF) annotation and AF annotation with external population callsets;
-
-- **Module 09 (in development)** Visualization, including scripts that generates IGV screenshots and rd plots.
-
-- Additional modules to be added: de novo and mosaic scripts
-
-
-## Pipeline Parameters
-
-Several inputs are shared across different modules of the pipeline, which are explained in this section.
-
-#### `ped_file`
-
-A pedigree file describing the familial relationships between the samples in the cohort.
-The file needs to be in the
-[PED format](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format).
-Updated with [EvidenceQC](./eqc) sex assignments, including
-`sex = 0` for sex aneuploidies;
-genotypes on chrX and chrY for samples with `sex = 0` in the PED file will be set to
-`./.` and these samples will be excluded from sex-specific training steps.
+For details on running GATK-SV on Terra, refer to the [Execution](/docs/execution/joint#instructions) section.
diff --git a/website/docs/modules/join_raw_calls.md b/website/docs/modules/join_raw_calls.md
new file mode 100644
index 000000000..55b563a24
--- /dev/null
+++ b/website/docs/modules/join_raw_calls.md
@@ -0,0 +1,54 @@
+---
+title: JoinRawCalls
+description: Clusters unfiltered variants across batches
+sidebar_position: 17
+slug: jrc
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/JoinRawCalls.wdl)
+
+This module clusters raw unfiltered variants from [ClusterBatch](./cb) across all batches. Concordance between these
+genotypes and the joint call set usually can be indicative of variant quality and is used downstream for genotype
+filtering.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cb: ClusterBatch
+ jrc: JoinRawCalls
+ svc: SVConcordance
+ cb --> jrc
+ jrc --> svc
+
+ class jrc thisModule
+ class cb inModules
+ class svc outModules
+```
+
+### Inputs
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. The guidelines outlined in the
+[sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `clustered_*_vcfs`
+Array of clustered VCFs for all batches, generated in [ClusterBatch](./cb#clustered__vcf).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+### Outputs
+
+#### `joined_raw_calls_vcf`
+VCF containing all raw calls in the cohort.
+
+#### `ploidy_table`
+TSV of contig ploidies for all samples, assuming diploid autosome and sex assignments from the [ped_file](#ped_file).
diff --git a/website/docs/modules/main_vcf_qc.md b/website/docs/modules/main_vcf_qc.md
new file mode 100644
index 000000000..d362765b4
--- /dev/null
+++ b/website/docs/modules/main_vcf_qc.md
@@ -0,0 +1,228 @@
+---
+title: MainVcfQC
+description: VCF QC plotting
+sidebar_position: 21
+slug: mvqc
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MainVcfQC.wdl)
+
+Creates plots for call set analysis and quality control (QC). This module is run by default with the
+[FilterGenotypes](./fg) module. Note, however, that the module is a stand-alone workflow and can be used with any batch-
+or cohort-level (i.e. multi-sample) SV VCF, including outputs from [ClusterBatch](./cb) and onward.
+
+Users can optionally provide external benchmarking datasets with the
+[site_level_comparison_datasets](#optional-site_level_comparison_datasets) and
+[sample_level_comparison_datasets](#optional-sample_level_comparison_datasets) parameters.
+
+The following sections provide guidance on interpreting these plots for VCFs that have been run with the recommended
+pipeline through [FilterGenotypes](./fg).
+
+### Output structure
+
+The tarball output has the following directory structure:
+
+- `/plots/main_plots` : main QC plots, described in detail [below](#plots-description).
+- `/plots/supplementary_plots` : comprehensive set of QC plots with greater detail, some of which are used as panels
+ in `main_plots`.
+- `/data` : various datafiles from the QC process such as family structures, a list of the samples
+ used for plotting, variant statistics, and lists of overlapping variants against any external call sets provided.
+
+### Plots description
+
+This section summarizes the most important plots used for overall call set QC found in the `/plots/main_plots`
+directory.
+
+Example plots are provided from a high-quality dataset consisting of 161 samples from the 1000 Genomes Project aligned
+with DRAGEN v3.7.8. Note that QC metrics and distributions will vary among data sets, particularly as sample size
+increases, and these examples are intended to provide a simple baseline for acceptable call set quality. Please see
+the [Recommendations](#recommendations) section for prescribed quality criteria.
+
+:::note
+The following plots are of variants passing all filters (i.e. with the `FILTER` status set to `PASS`). This is the
+default behavior of the QC plots generated in [FilterGenotypes](./fg).
+
+When running `MainVcfQc` as a standalone workflow, users may set the
+[bcftools_preprocessing_options](#optional--bcftools_preprocessing_options) argument to limit plotted variants based on `FILTER` status.
+For example, to limit to `PASS` variants for a VCF generated from [FilterGenotypes](./fg) use:
+```
+"bcftools_preprocessing_options": "-i 'FILTER~\"PASS\""
+```
+:::
+
+#### SV per genome
+
+Shows per-genome SV count distributions broken down by SV type. The top row is a site-level analysis, and
+the bottom row provides counts of individual alleles. Distributions of genotype quality (`GQ`), SV size, and
+frequency are also shown.
+
+
+
+#### SV counts
+
+Summarizes total variant counts by type, frequency, and size.
+
+
+
+#### Genotype distributions
+
+Distributions of genotypes across the call set.
+
+The left panel plots carrier frequency against allele frequency and can be used to visualize genotyping bias. In this
+plot, the rolling mean should not lie on the lower or upper extremes (all het. or all hom.).
+
+The middle panel plots the Hardy-Weinberg (HW) distribution of the cohort and provides estimates for the proportions of
+variants in HW equilibrium (or nominally in HW equilibrium before a multiple testing correction). The right-hand
+plots stratify HW by SV size.
+
+:::note
+The horizontal line bisecting the vertical axis in the AF vs. carrier frequency plot was fixed in.
+[#625](https://github.com/broadinstitute/gatk-sv/pull/625)).
+:::
+
+
+
+#### Size distributions
+
+Contains plots of SV length stratified by class and frequency.
+
+
+
+#### Frequency distributions
+
+Contains plots of allele frequency stratified by class and size.
+
+
+
+#### Optional SV trio inheritance
+
+Inheritance analysis of sites and alleles, including *de novo* rate distributions by SV class, size, frequency, and
+genotype quality. This plot is only generated if the cohort contains trios defined in the [PED file](#optional-ped_file).
+
+
+
+#### Optional Call set benchmarking
+
+This plot will be generated for each external call set provided. The left-hand panels show distributions of overlapping
+variants in the input VCF by SV class, size, and frequency. Colors depict proportions of variants meeting each overlap
+criterion. The right-hand panels show distributions among variants overlapping between the input and external call sets
+by SV class, size, and frequency.
+
+:::note
+Note the horizontal line bisecting the vertical axis in the AF vs. AF plot has been fixed in
+[#625](https://github.com/broadinstitute/gatk-sv/pull/625)).
+:::
+
+
+
+
+### Recommendations
+
+We suggest users observe the following basic criteria to assess the overall quality of the final call set:
+
+* Number of PASS variants per genome (which excludes `BND`) between 7,000 and 11,000.
+* Relative overall proportions of SV types similar to those shown above.
+* Similar size and frequency distributions to those shown above.
+* At least 70% of variants in HW equilibrium. Note that this may be lower depending on how well
+ the underlying cohort population structure adheres to the assumptions of the HW model. Note that HW equilibrium
+ usually improves after genotype filtering.
+* Strong allele frequency correlation against an external benchmarking dataset such as gnomAD-v2.
+* Low allele *de novo* inheritance rate (if trios are present), typically below 10%.
+
+These are intended as general guidelines and not strict limits on call set quality.
+
+### Inputs
+
+#### `vcfs`
+Array of one or more input VCFs. Each VCF must be indexed. This input is structured as an `Array` type for convenience
+when VCFs are sharded by contig. All VCFs in the array are concatenated before processing.
+
+#### Optional `vcf_format_has_cn`
+Default: `true`. This must be set to `false` for VCFs from modules before [CleanVcf](./cvcf) (i.e. without `CN` fields).
+
+#### Optional `bcftools_preprocessing_options`
+Additional arguments to pass to [bcftools view](https://samtools.github.io/bcftools/bcftools.html) before running
+the VCF through QC tools.
+
+#### Optional `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+If provided and there are trios present then [SV trio plots](#optional-sv-trio-inheritance) will be generated.
+
+#### Optional `list_of_samples_to_include`
+If provided, the input VCF(s) will be subset to samples in the list.
+
+#### Optional `sample_renaming_tsv`
+File with mapping to rename per-sample benchmark sample IDs for compatibility with cohort (see
+[here](/docs/resources#hgsv_byrska_bishop_sample_renaming_tsv) for example).
+
+#### Optional `max_trios`
+Default: `1000`. Upper limit on the number of trios to use for inheritance analysis.
+
+#### `prefix`
+Output prefix, such as cohort name. May be alphanumeric with underscores.
+
+#### `sv_per_shard`
+Records per shard for parallel processing. This parameter may be reduced if the workflow is running slowly.
+
+#### `samples_per_shard`
+Samples per shard for per-sample QC processing. This parameter may be reduced if the workflow is running slowly. Only
+has an effect when [do_per_sample_qc](#optional-do_per_sample_qc) is enabled.
+
+#### Optional `do_per_sample_qc`
+Default: `true`. If enabled, performs per-sample SV count plots, family analysis, and site-level benchmarking.
+
+#### Optional `site_level_comparison_datasets`
+Array of two-element arrays, one per dataset, each a tuple containing `[prefix, bed_uri_path]`, where the latter is
+a directory containing multiple bed files containing the call set stratified by subpopulation. For example:
+
+```
+["gnomAD_v2_Collins", "gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins"]
+```
+
+where the bucket directory contains the following files:
+
+```
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.AFR.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.ALL.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.AMR.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.EAS.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.EUR.bed.gz
+gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/gnomAD_v2_Collins/gnomAD_v2_Collins.SV.OTH.bed.gz
+```
+
+Users can examine the above files as an example of how to format custom benchmarking datasets.
+
+See the `*_site_level_benchmarking_dataset` entries in the [Resource files](/docs/resources) section for available
+benchmarking call sets.
+
+#### Optional `sample_level_comparison_datasets`
+Array of two-element arrays, one per dataset, each a tuple containing `[prefix, tarball_uri]`, where the latter is
+a tarball containing sample-level benchmarking data. For example:
+
+```
+[["HGSV_ByrskaBishop", "gs://gatk-sv-resources-public/hg38/v0/sv-resources/resources/v1/HGSV_ByrskaBishop_GATKSV_perSample.tar.gz"]]
+```
+
+Users can examine the above file as an example of how to format custom benchmarking datasets.
+
+See the `*_sample_level_benchmarking_dataset` entries in the [Resource files](/docs/resources#benchmarking-datasets)
+section for available benchmarking call sets.
+
+#### Optional `random_seed`
+Default: `0`. Random seed for sample subsetting in external call set comparisons.
+
+#### Optional `max_gq`
+Default: `99`. Max value to define range for `GQ` plotting. For modules prior to `CleanVcf`, use `999`.
+
+#### Optional `downsample_qc_per_sample`
+Default: `1000`. Number of samples to subset to for per-sample QC.
+
+### Outputs
+
+#### `sv_vcf_qc_output`
+Tarball of QC plots and data tables.
+
+#### `vcf2bed_output`
+Bed file containing all input variants.
diff --git a/website/docs/modules/make_cohort_vcf.md b/website/docs/modules/make_cohort_vcf.md
deleted file mode 100644
index d16e22acf..000000000
--- a/website/docs/modules/make_cohort_vcf.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-title: MakeCohortVcf
-description: Make Cohort VCF
-sidebar_position: 11
-slug: cvcf
----
-
-Combines variants across multiple batches, resolves complex variants,
-re-genotypes, and performs final VCF clean-up.
-
-### Prerequisites
-
-- GenotypeBatch
-- (Optional) RegenotypeCNVs
-
-### Inputs
-
-- RD, PE and SR file URIs (GatherBatchEvidence)
-- Batch filtered PED file URIs (FilterBatch)
-- Genotyped PESR VCF URIs (GenotypeBatch)
-- Genotyped depth VCF URIs (GenotypeBatch or RegenotypeCNVs)
-- SR pass variant file URIs (GenotypeBatch)
-- SR fail variant file URIs (GenotypeBatch)
-- Genotyping cutoff file URIs (GenotypeBatch)
-- Batch IDs
-- Sample ID list URIs
-
-### Outputs
-
-- Finalized "cleaned" VCF and QC plots
\ No newline at end of file
diff --git a/website/docs/modules/merge_batch_sites.md b/website/docs/modules/merge_batch_sites.md
index 6bfe43789..c1f789c09 100644
--- a/website/docs/modules/merge_batch_sites.md
+++ b/website/docs/modules/merge_batch_sites.md
@@ -5,16 +5,66 @@ sidebar_position: 8
slug: msites
---
-Combines filtered variants across batches. The WDL can be found at: /wdl/MergeBatchSites.wdl.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MergeBatchSites.wdl)
-### Prerequisites
-- Filter Batch
+Merges variants across batches. Variants are merged only if the following attributes match exactly:
+
+- Contig
+- Start position
+- End position (`END` field)
+- SV type (`SVTYPE` field)
+- SV length (`SVLEN` field, if available)
+- Strandedness (`STRANDS` field, if available)
+- Second contig (`CHR2` field, if available)
+- Second end (`END2` field, if available)
+
+This is a "cohort-level" workflow, meaning that is aggregates data across all batches. This is in contrast to all previous
+modules, which are sample- or batch-level. Note that this workflow should still be run on cohorts consisting of
+a single batch.
+
+:::info
+Terra users must configure a "sample_set_set" in their data table before running this module. See the [Execution
+section on MergeBatchSites](/docs/execution/joint#09-mergebatchsites) for futher instructions.
+:::
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ fb: FilterBatch
+ mbs: MergeBatchSites
+ gb: GenotypeBatch
+ fb --> mbs
+ mbs --> gb
+
+ class mbs thisModule
+ class fb inModules
+ class gb outModules
+```
### Inputs
-- List of filtered PESR VCFs (FilterBatch)
-- List of filtered depth VCFs (FilterBatch)
+#### `cohort`
+An identifier for the cohort. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids)
+section apply here.
+
+#### `depth_vcfs`
+Array of filtered depth VCFs across batches generated in [FilterBatch](./fb#filtered_depth_vcf).
+
+#### `pesr_vcfs`
+Array of filtered depth VCFs across batches generated in [FilterBatch](./fb#filtered_pesr_vcf).
### Outputs
-- Combined cohort PESR and depth VCFs
+#### `cohort_pesr_vcf`
+Merged PE/SR caller VCF.
+
+#### `cohort_depth_vcf`
+Merged depth caller VCF.
diff --git a/website/docs/modules/refine_cpx.md b/website/docs/modules/refine_cpx.md
new file mode 100644
index 000000000..6ed5e0f41
--- /dev/null
+++ b/website/docs/modules/refine_cpx.md
@@ -0,0 +1,84 @@
+---
+title: RefineComplexVariants
+description: Refines and filters complex variants
+sidebar_position: 15
+slug: refcv
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/RefineComplexVariants.wdl)
+
+Refines complex SVs and translocations and filters based on discordant read pair and read depth evidence reassessment.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cvcf: CleanVcf
+ refcv: RefineComplexVariants
+ amvf: ApplyManualVariantFilter
+
+ cvcf --> refcv
+ refcv --> amvf
+
+ class refcv thisModule
+ class cvcf inModules
+ class amvf outModules
+```
+
+### Inputs
+
+:::info
+All array inputs of batch data must match in order. For example, the order of the `batch_name_list` array should match
+that of `batch_sample_lists`, `PE_metrics`, etc.
+:::
+
+#### `vcf`
+Input vcf, generated in [CleanVcf](./cvcf#cleaned_vcf).
+
+#### `prefix`
+Prefix for output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `batch_name_list`
+Array of batch names. These should be the same batch names used in [GatherBatchEvidence](./gbe#batch).
+
+#### `batch_sample_lists`
+Array of sample ID lists for all batches, generated in [FilterBatch](./fb#batch_samples_postoutlierexclusion). Order must match [batch_name_list](#batch_name_list).
+
+#### `PE_metrics`
+Array of PE metrics files for all batches, generated in [GatherBatchEvidence](./gbe#merged_pe). Order must match [batch_name_list](#batch_name_list).
+
+#### `Depth_DEL_beds`, `Depth_DUP_beds`
+Arrays of raw DEL and DUP depth calls for all batches, generated in [GatherBatchEvidence](./gbe#merged_dels-merged_dups). Order must match [batch_name_list](#batch_name_list).
+
+#### `n_per_split`
+Shard size for parallel computations. Decreasing this parameter may help reduce run time.
+
+#### Optional `min_pe_cpx`
+Default: `3`. Minimum PE read count for complex variants (CPX).
+
+#### Optional `min_pe_ctx`
+Default: `3`. Minimum PE read count for translocations (CTX).
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `cpx_refined_vcf`
+Output VCF.
+
+#### `cpx_evidences`
+Supplementary output table of complex variant evidence.
diff --git a/website/docs/modules/regenotype_cnvs.md b/website/docs/modules/regenotype_cnvs.md
index cb5356b7e..b05dc2afe 100644
--- a/website/docs/modules/regenotype_cnvs.md
+++ b/website/docs/modules/regenotype_cnvs.md
@@ -1,24 +1,79 @@
---
-title: ReGenotypeCNVs
+title: RegenotypeCNVs
description: Regenotype CNVs
sidebar_position: 10
slug: rgcnvs
---
-Re-genotypes probable mosaic variants across multiple batches.
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/RegenotypeCNVs.wdl)
-### Prerequisites
-- Genotype batch
+Re-genotypes probable mosaic variants across multiple batches. This is a "cohort-level" workflow that operates on
+all batches.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ gb: GenotypeBatch
+ rgc: RegenotypeCNVs
+ cb: CombineBatches
+ gb --> rgc
+ rgc --> cb
+
+ class rgc thisModule
+ class gb inModules
+ class cb outModules
+```
### Inputs
-- Per-sample median coverage estimates (GatherBatchEvidence)
-- Pre-genotyping depth VCFs (FilterBatch)
-- Batch PED files (FilterBatch)
-- Cohort depth VCF (MergeBatchSites)
-- Genotyped depth VCFs (GenotypeBatch)
-- Genotyped depth RD cutoffs file (GenotypeBatch)
+:::info
+All array inputs of batch data must match in order. For example, the order of the `batches` array should match that of
+`depth_vcfs`, `batch_depth_vcfs`, etc.
+:::
+
+#### `depth_vcfs`
+Array of genotyped depth caller variants for all batches, generated in [GenotypeBatch](./gb#genotyped_depth_vcf).
+
+#### `cohort_depth_vcf`
+Merged depth caller variants for the cohort, generated in [MergeBatchSites](./msites#cohort_depth_vcf).
+
+#### `batch_depth_vcfs`
+Array of filtered depth caller variants for all batches, generated in [FilterBatch](./fb#filtered_depth_vcf). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `coveragefiles`
+Array of merged RD evidence files for all batches from [GatherBatchEvidence](./gbe#merged_bincov). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `medianfiles`
+Array of median coverage tables for all batches from [GatherBatchEvidence](./gbe#median_cov). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `RD_depth_sepcutoffs`
+Array of "depth_depth" genotype cutoff files (depth evidence for depth-based calls) generated in
+[GenotypeBatch](./gb#trained_genotype___sepcutoff). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `n_per_split`
+Records per shard when scattering variants. Decrease to increase parallelism if the workflow is running slowly.
+
+#### `n_RD_genotype_bins`
+Number of depth genotyping bins. Most users should leave this at the default value.
+
+#### `batches`
+Array of batch identifiers. Should match the name used in [GatherBatchEvidence](./gbe#batch). Order must match that of [depth_vcfs](#depth_vcfs).
+
+#### `cohort`
+Cohort name. May be alphanumeric with underscores.
+
+#### `regeno_coverage_medians`
+Array of regenotyping metrics generated in [GenotypeBatch](./gb#regeno_coverage_medians).
### Outputs
-- Re-genotyped depth VCFs.
+#### `regenotyped_depth_vcfs`
+Array of batch depth VCFs after regenotyping.
diff --git a/website/docs/modules/resolve_complex.md b/website/docs/modules/resolve_complex.md
new file mode 100644
index 000000000..5121e1a12
--- /dev/null
+++ b/website/docs/modules/resolve_complex.md
@@ -0,0 +1,89 @@
+---
+title: ResolveComplexVariants
+description: Complex SV discovery
+sidebar_position: 12
+slug: rcv
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/ResolveComplexVariants.wdl)
+
+Identifies multi-breakpoint complex variants, which are annotated with the `CPX` value in the `SVTYPE` field. These
+variants are putative, as read depth evidence is not assessed at this stage.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cb: CombineBatches
+ rcv: ResolveComplexVariants
+ gcv: GenotypeComplexVariants
+ cb --> rcv
+ rcv --> gcv
+
+ class rcv thisModule
+ class cb inModules
+ class gcv outModules
+```
+
+### Inputs
+
+:::info
+Some inputs of batch data must match in order. Specifically, the order of the `disc_files` array should match that of
+`rf_cutoff_files`.
+:::
+
+#### `cohort_name`
+Cohort name. The guidelines outlined in the [sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### Optional `merge_vcfs`
+Default: `false`. If true, merge contig-sharded VCFs into one genome-wide VCF. This may be used for convenience but cannot be used with
+downstream workflows.
+
+#### `cluster_vcfs`
+Array of contig-sharded VCFs, generated in [CombineBatches](./cmb#combined_vcfs).
+
+#### `cluster_bothside_pass_lists`
+Array of variant lists with bothside SR support for all batches, generated in [CombineBatches](./cmb#cluster_bothside_pass_lists).
+
+#### `cluster_background_fail_lists`
+Array of variant lists with low SR signal-to-noise ratio for all batches, generated in [CombineBatches](./cmb#cluster_background_fail_lists).
+
+#### `disc_files`
+Array of PE evidence files for all batches from [GatherBatchEvidence](./gbe#merged_pe).
+
+#### `rf_cutoffs`
+Array of batch genotyping cutoff files trained with the random forest filtering model from [FilterBatch](./fb#cutoffs).
+Must match the order of [disc_files](#disc_files).
+
+#### Optional `use_hail`
+Default: `false`. Use Hail for VCF concatenation. This should only be used for projects with over 50k samples. If enabled, the
+[gcs_project](#optional-gcs_project) must also be provided. Does not work on Terra.
+
+#### Optional `gcs_project`
+Google Cloud project ID. Required only if enabling [use_hail](#optional-use_hail).
+
+### Outputs
+
+#### `complex_resolve_vcfs`
+Array of contig-sharded VCFs containing putative complex variants.
+
+#### `complex_resolve_bothside_pass_list`
+Array of contig-sharded bothside SR support variant lists.
+
+#### `complex_resolve_background_fail_list`
+Array of contig-sharded high SR background variant lists.
+
+#### `breakpoint_overlap_dropped_record_vcfs`
+Variants dropped due to exact overlap with another's breakpoint.
+
+#### `complex_resolve_merged_vcf`
+Genome-wide output VCF. Only generated if using [merge_vcfs](#optional--merge_vcfs).
diff --git a/website/docs/modules/train_gcnv.md b/website/docs/modules/train_gcnv.md
index 45c7b3cac..abd45ba8e 100644
--- a/website/docs/modules/train_gcnv.md
+++ b/website/docs/modules/train_gcnv.md
@@ -7,40 +7,33 @@ slug: gcnv
import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/TrainGCNV.wdl)
+
[GATK-gCNV](https://www.nature.com/articles/s41588-023-01449-0)
is a method for detecting rare germline copy number variants (CNVs)
from short-read sequencing read-depth information.
-The [TrainGCNV](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/TrainGCNV.wdl)
-module trains a gCNV model for use in the [GatherBatchEvidence](./gbe) workflow.
-The upstream and downstream dependencies of the TrainGCNV module are illustrated in the following diagram.
-
+The `TrainGCNV` module trains a gCNV model for use in the [GatherBatchEvidence](./gbe) workflow.
The samples used for training should be homogeneous (concerning sequencing platform,
-coverage, library preparation, etc.) and similar
-to the samples on which the model will be applied in terms of sample type,
-library preparation protocol, sequencer, sequencing center, and etc.
-
+coverage, library preparation, etc.) and similar to the samples on which the model will be applied.
For small, relatively homogeneous cohorts, a single gCNV model is usually sufficient.
However, for larger cohorts, especially those with multiple data sources,
we recommend training a separate model for each batch or group of batches (see
-[batching section](/docs/run/joint#batching) for details).
+[batching section](/docs/execution/joint#batching) for details).
The model can be trained on all or a subset of the samples to which it will be applied.
A subset of 100 randomly selected samples from the batch is a reasonable
input size for training the model; when the `n_samples_subsample` input is provided,
the `TrainGCNV` workflow can automatically perform this random selection.
-The following diagram illustrates the upstream and downstream workflows of the `TrainGCNV` workflow
-in the recommended invocation order. You may refer to
-[this diagram](https://github.com/broadinstitute/gatk-sv/blob/main/terra_pipeline_diagram.jpg)
-for the overall recommended invocation order.
+The following diagram illustrates the recommended invocation order:
```mermaid
stateDiagram
direction LR
- classDef inModules stroke-width:0px,fill:#00509d,color:#caf0f8
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
@@ -58,49 +51,59 @@ stateDiagram
## Inputs
-This section provides a brief description on the _required_ inputs of the TrainGCNV workflow.
-For a description on the _optional_ inputs and their default values, you may refer to the
-[source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/TrainGCNV.wdl) of the TrainGCNV workflow.
-Additionally, the majority of the optional inputs of the workflow map to the optional arguments of the
-tool the workflow uses, `GATK GermlineCNVCaller`; hence, you may refer to the
+The majority of the optional inputs of the workflow map to the optional arguments of the
+tool the workflow uses, `GATK-GermlineCNVCaller`; hence, you may refer to the
[documentation](https://gatk.broadinstitute.org/hc/en-us/articles/360040097712-GermlineCNVCaller)
-of the tool for a description on these optional inputs.
+of the tool for a description on these optional inputs. We recommend that most users use the defaults.
+
+:::info
+All array inputs of sample data must match in order. For example, the order of the `samples` array should match that
+of the `count_files` array.
+:::
#### `samples`
-A list of sample IDs.
-The order of IDs in this list should match the order of files in `count_files`.
+Sample IDs
#### `count_files`
-A list of per-sample coverage counts generated in the [GatherSampleEvidence](./gse#outputs) workflow.
+Per-sample binned read counts (`*.rd.txt.gz`) generated in the [GatherSampleEvidence](./gse#outputs) workflow.
-#### `contig_ploidy_priors`
-A tabular file with ploidy prior probability per contig.
-You may find the link to this input from
-[this reference](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json)
-and a description to the file format
-[here](https://gatk.broadinstitute.org/hc/en-us/articles/360037224772-DetermineGermlineContigPloidy).
+#### Optional `n_samples_subsample`, `sample_ids_training_subset`
+Provide one of these inputs to subset the input batch. `n_samples_subsample` will randomly subset, while
+`sample_ids_training_subset` is for defining a predetermined subset. These options are provided for convenience in Terra.
+## Outputs
-#### `reference_fasta`
-`reference_fasta`, `reference_index`, `reference_dict` are respectively the
-reference genome sequence in the FASTA format, its index file, and a corresponding
-[dictionary file](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format).
-You may find links to these files from
-[this reference](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json).
+#### `cohort_contig_ploidy_model_tar`
+Contig ploidy model tarball.
+#### `cohort_gcnv_model_tars`
+CNV model tarballs scattered across genomic intervals.
-## Outputs
+#### `cohort_contig_ploidy_calls_tar`
+Contig ploidy calls for the submitted batch.
-#### Optional `annotated_intervals` {#annotated-intervals}
+#### `cohort_gcnv_calls_tars`
+CNV call tarballs scattered by sample and genomic region prior to segmentation.
+#### `cohort_genotyped_segments_vcfs`
+Single-sample VCFs of CNV calls for the submitted batch.
+
+#### `cohort_gcnv_tracking_tars`
+Convergence tracking logs.
+
+#### `cohort_genotyped_intervals_vcfs`
+Single-sample VCFs for the submitted batch containing per-interval genotypes prior to segmentation.
+
+#### `cohort_denoised_copy_ratios`
+TSV files containing denoised copy ratios in each sample.
+
+#### Optional `annotated_intervals`
The count files from [GatherSampleEvidence](./gse) with adjacent intervals combined into
locus-sorted `DepthEvidence` files using `GATK CondenseDepthEvidence` tool, which are
annotated with GC content, mappability, and segmental-duplication content using
-[`GATK AnnotateIntervals`](https://gatk.broadinstitute.org/hc/en-us/articles/360041416652-AnnotateIntervals)
-tool. This output is generated if the optional input `do_explicit_gc_correction` is set to `True`.
-
-#### Optional `filtered_intervals_cnv` {#filtered-intervals-cnv}
-
-#### Optional `cohort_contig_ploidy_model_tar` {#contig-ploidy-model-tarball}
+[`GATK-AnnotateIntervals`](https://gatk.broadinstitute.org/hc/en-us/articles/360041416652-AnnotateIntervals)
+tool. This output is generated if `do_explicit_gc_correction` is set to `True`. Disabled by default.
-#### Optional `cohort_gcnv_model_tars` {#model-tarballs}
+#### Optional `filtered_intervals_cnv`, `filtered_intervals_ploidy`
+Intervals of read count bins to be used for CNV and ploidy calling after filtering for problematic regions (e.g.
+high GC content). This output is generated if `filter_intervals` is set to `True`. Enabled by default.
diff --git a/website/docs/modules/visualization.md b/website/docs/modules/visualization.md
deleted file mode 100644
index c6c71bf65..000000000
--- a/website/docs/modules/visualization.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-title: Visualization
-description: Visualization (work in progress)
-sidebar_position: 14
-slug: viz
----
-
-Visualize SVs with IGV screenshots and read depth plots.
-
-Visualization methods include:
-
-- RD Visualization - generate RD plots across all samples, ideal for visualizing large CNVs.
-- IGV Visualization - generate IGV plots of each SV for individual sample, ideal for visualizing de novo small SVs.
-- Module09.visualize.wdl - generate RD plots and IGV plots, and combine them for easy review.
diff --git a/website/docs/modules/visualize_cnvs.md b/website/docs/modules/visualize_cnvs.md
new file mode 100644
index 000000000..c0e792117
--- /dev/null
+++ b/website/docs/modules/visualize_cnvs.md
@@ -0,0 +1,51 @@
+---
+title: VisualizeCnvs
+description: Visualize CNVs
+sidebar_position: 22
+slug: viz
+---
+
+Generates plots of read depth across all samples. This is useful for visualizing large deletions and duplications
+(at least 5kbp).
+
+This module is not a part of the core pipeline but may be used to investigate variants of interest.
+
+### Inputs
+
+:::info
+All array inputs of batch data must match in order. In particular, the ordering of `median_files` and `rd_files` must
+be the same.
+:::
+
+#### `vcf_or_bed`
+VCF or bed file containing variants to plot. All variants will be automatically subsetted to DEL and DUP types subject
+to the [min_size](#min_size) constraint. VCF files must end in `.vcf.gz` and bed files must end in either `.bed` or
+`.bed.gz`. Bed files must contain columns: `chrom,start,end,name,svtype,samples`.
+
+#### `prefix`
+Output prefix, such as cohort name. May be alphanumeric with underscores.
+
+#### `median_files`
+Array of median coverage files for all batches in the input variants, generated in [GatherBatchEvidence](./gbe#median_cov).
+
+#### `rd_files`
+Array of RD evidence files for all batches in the input variants, generated in [GatherBatchEvidence](./gbe#merged_bincov).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+#### `min_size`
+Minimum size in bases of variants to plot.
+
+#### `flags`
+Additional flags to pass to the [RdTest plotting script](https://github.com/broadinstitute/gatk-sv/blob/main/src/RdTest/RdTest.R).
+
+:::warning
+Due to a bug, the `flags` parameter must contain `-s 999999999` in order to properly plot variants over 1 Mb.
+:::
+
+### Outputs
+
+#### `rdtest_plots`
+Tarball containing output plots.
+
diff --git a/website/docs/references.md b/website/docs/references.md
new file mode 100644
index 000000000..c30d1da62
--- /dev/null
+++ b/website/docs/references.md
@@ -0,0 +1,30 @@
+---
+title: Citation and references
+description: Citation and references
+sidebar_position: 11
+---
+
+### How to cite
+
+To credit GATK-SV, please cite the following publication:
+
+- [Collins RL et al. A structural variation reference for medical and population genetics. Nature. 2020 May;581(7809):444-451.](https://doi.org/10.1038/s41586-020-2287-8)
+
+### Additional references
+
+The following is a selected list of studies that have used GATK-SV:
+
+- [Belyeu JR et al. De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2,396 families. Am J Hum Genet. 2021 Apr 1;108(4):597-607.](https://doi.org/10.1016/j.ajhg.2021.02.012)
+- [Billingsley KJ et al. Genome-Wide Analysis of Structural Variants in Parkinson Disease. Ann Neurol. 2023 May;93(5):1012-1022.](https://doi.org/10.1002/ana.26608)
+- [Byrska-Bishop M et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell. 2022 Sep 1;185(18):3426-3440.e19.](https://doi.org/10.1016/j.cell.2022.08.004)
+- [Diaz Perez KK et al. Rare variants found in clinical gene panels illuminate the genetic and allelic architecture of orofacial clefting. Genet Med. 2023 Oct;25(10):100918.](https://doi.org/10.1016/j.gim.2023.100918)
+- [Gillani R et al. Rare germline structural variants increase risk for pediatric solid tumors. bioRxiv [Preprint]. 2024 Apr 29:2024.04.27.591484.](https://doi.org/10.1101/2024.04.27.591484)
+- [Han L et al. Functional annotation of rare structural variation in the human brain. Nat Commun. 2020 Jun 12;11(1):2990.](https://doi.org/10.1038/s41467-020-16736-1)
+- [Jurgens JA et al. Expanding the genetics and phenotypes of ocular congenital cranial dysinnervation disorders. Genet Med. 2024 Jul 17:101216.](https://doi.org/10.1016/j.gim.2024.101216)
+- [Kaivola K et al. Genome-wide structural variant analysis identifies risk loci for non-Alzheimer's dementias. Cell Genom. 2023 May 4;3(6):100316.](https://doi.org/10.1016/j.xgen.2023.100316)
+- [Koenig Z et al. A harmonized public resource of deeply sequenced diverse human genomes. Genome Res. 2024 Jun 25;34(5):796-809.](https://doi.org/10.1101/gr.278378.123)
+- [Lee AS et al. A cell type-aware framework for nominating non-coding variants in Mendelian regulatory disorders. Nat Commun. 2024 Sep 27;15(1):8268.](https://doi.org/10.1038/s41467-024-52463-7)
+- [Lowther C et al. Systematic evaluation of genome sequencing for the diagnostic assessment of autism spectrum disorder and fetal structural anomalies. Am J Hum Genet. 2023 Sep 7;110(9):1454-1469.](https://doi.org/10.1016/j.ajhg.2023.07.010)
+- [Werling DM et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet. 2018 Apr 26;50(5):727-736.](https://doi.org/10.1038/s41588-018-0107-y)
+- [Wojcik MH et al. Genome Sequencing for Diagnosing Rare Diseases. N Engl J Med. 2024 Jun 6;390(21):1985-1997.](https://doi.org/10.1056/nejmoa2314761)
+- [Zhao B et al. A neurodevelopmental disorder caused by a novel de novo SVA insertion in exon 13 of the SRCAP gene. Eur J Hum Genet. 2022 Sep;30(9):1083-1087.](https://doi.org/10.1038/s41431-022-01137-3)
diff --git a/website/docs/resources.md b/website/docs/resources.md
new file mode 100644
index 000000000..c27a13d5a
--- /dev/null
+++ b/website/docs/resources.md
@@ -0,0 +1,174 @@
+---
+title: Resource files
+description: Resource files manifest
+sidebar_position: 7
+---
+
+This page contains descriptions of common resource files used in the pipeline. All required files are publicly available
+in Google Cloud Storage Buckets. URIs for these files are available in the [hg38 resources json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/resources_hg38.json).
+
+:::info
+Some resources contain sensitive data and are stored in a secure bucket for development purposes. These files are not
+required to run GATK-SV.
+:::
+
+### Reference resources
+
+#### allosome_file
+Reference [fasta index file](https://www.htslib.org/doc/faidx.html) containing only allosomal contigs.
+
+#### autosome_file
+Reference [fasta index file](https://www.htslib.org/doc/faidx.html) containing only autosomal contigs.
+
+#### bin_exclude
+[Block-compressed](https://www.htslib.org/doc/bgzip.html) [bed file](https://genome.ucsc.edu/FAQ/FAQformat.html#format1)
+of intervals to exclude in the call set.
+
+#### cnmops_exclude_list
+Plain text bed file of N-masked regions in the reference.
+
+#### contig_ploidy_priors
+Plain text TSV file of prior probabilities contig ploidies used by GATK-gCNV.
+
+#### cytobands
+Block-compressed bed file of cytoband intervals.
+
+#### sd_locs_vcf
+Plain text VCF of SNP sites at which to collect site depth (SD) evidence.
+
+#### depth_exclude_list
+Block-compressed bed file of intervals over which to exclude overlapping depth-only calls.
+
+#### empty_file
+Empty file; used to satisfy some workflow code paths.
+
+#### exclude_intervals_for_gcnv_filter_intervals
+Plain text bed file of intervals to exclude from GATK-gCNV.
+
+#### external_af_ref_bed
+Block-compressed bed file of SV sites for external allele frequency annotation.
+
+#### genome_file
+Tab-delimited table of primary contigs with contig name in the first column and length in the second column.
+
+#### manta_region_bed
+Block-compressed bed file of intervals to call with Manta.
+
+#### mei_bed
+Block-compressed bed file of mobile element insertions in the reference genome.
+
+#### melt_std_vcf_header
+Text file containing the VCF header for raw MELT calls.
+
+#### noncoding_bed
+Plain text bed file of non-coding elements in the reference genome.
+
+#### par_bed
+Plain text bed file of pseudoautosomal regions.
+
+#### pesr_exclude_list
+Block-compressed bed file of intervals for filtering calls. Variants generated with non-CNV tools (Manta, MELT,
+Scramble, Wham) that have either end in any of these intervals are hard-filtered.
+
+#### preprocessed_intervals
+Intervals for read count collection and CNV calling.
+
+#### primary_contigs_fai
+Reference [fasta index file](https://www.htslib.org/doc/faidx.html) only containing the primary contigs, i.e. `chr1`, ...,
+`chr22`, `chrX`, and `chrY`.
+
+#### primary_contigs_list
+Text file of primary contig names.
+
+#### contigs_header
+Plain text VCF header section of primary contig sequences.
+
+#### protein_coding_gtf
+Protein coding sequence definitions for functional annotation in [General Transfer Format](https://www.ensembl.org/info/website/upload/gff.html).
+
+#### reference_dict
+Reference FASTA dictionary file (`*.dict`). See [this article](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format) for more information.
+
+#### reference_fasta
+Reference FASTA file (`*.fasta`). See [this article](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format) for more information.
+
+#### reference_index
+Reference FASTA index file (`*.fasta.fai`). See [this article](https://gatk.broadinstitute.org/hc/en-us/articles/360035531652-FASTA-Reference-genome-format) for more information.
+
+#### rmsk
+Block-compressed bed file of [RepeatMasker intervals](https://genome.ucsc.edu/cgi-bin/hgTrackUi?g=rmsk).
+
+#### segdups
+Block-compressed bed file of [segmental duplication intervals](https://genome.ucsc.edu/cgi-bin/hgTrackUi?g=genomicSuperDups).
+
+#### seed_cutoffs
+TSV of cutoff priors for genotyping.
+
+#### single_sample_qc_definitions
+TSV of recommended ranges for single-sample QC metrics.
+
+#### wgd_scoring_mask
+Plain text bed file of whole-genome dosage (WGD) score intervals over which to assess coverage bias.
+
+#### wham_include_list_bed_file
+Plain text bed file of intervals to call with Wham.
+
+#### aou_recalibrate_gq_model_file
+Genotype filtering model trained using data generated by All of Us.
+
+#### hgdp_recalibrate_gq_model_file
+Genotype filtering model trained using data generated from the Human Genome Diversity Project.
+
+#### recalibrate_gq_genome_tracks
+List of block-compressed bed files, each containing intervals from a separate genome track.
+
+### Benchmarking datasets
+
+#### ccdg_abel_site_level_benchmarking_dataset
+Benchmarking variant set from [Abel et al. 2020](https://doi.org/10.1038/s41586-020-2371-0).
+
+#### gnomad_v2_collins_sample_level_benchmarking_dataset
+Benchmarking genotypes from gnomAD-SV-v2, see [Collins et al. 2020](https://doi.org/10.1038/s41586-020-2287-8). **Not public data.**
+
+#### gnomad_v2_collins_site_level_benchmarking_dataset
+Benchmarking variant set from gnomAD-SV-v2, see [Collins et al. 2020](https://doi.org/10.1038/s41586-020-2287-8).
+
+#### hgsv_byrska_bishop_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Byrska-Bishop et al. 2022](https://doi.org/10.1016/j.cell.2022.08.004).
+
+#### hgsv_byrska_bishop_sample_renaming_tsv
+Sample renaming manifest for the Byrska-Bishop benchmarking genotypes.
+
+#### hgsv_byrska_bishop_site_level_benchmarking_dataset
+Benchmarking variant set from [Byrska-Bishop et al. 2022](https://doi.org/10.1016/j.cell.2022.08.004).
+
+#### hgsv_ebert_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Ebert et al. 2021](https://doi.org/10.1126/science.abf7117).
+
+#### ssc_belyeu_sample_level_benchmarking_dataset
+Benchmarking genotypes from the [Simons Simplex Collection](https://www.sfari.org/resource/simons-simplex-collection/), derived from [Belyeu et al. 2021](https://doi.org/10.1016/j.ajhg.2021.02.012). **Not public data.**
+
+#### ssc_belyeu_site_level_benchmarking_dataset
+Benchmarking variant set from the [Simons Simplex Collection](https://www.sfari.org/resource/simons-simplex-collection/), derived from [Belyeu et al. 2021](https://doi.org/10.1016/j.ajhg.2021.02.012). **Not public data.**
+
+#### ssc_sanders_sample_level_benchmarking_dataset
+Benchmarking genotypes from the [Simons Simplex Collection](https://www.sfari.org/resource/simons-simplex-collection/), derived from [Sanders et al. 2015](https://doi.org/10.1016/j.neuron.2015.09.016). **Not public data.**
+
+#### thousand_genomes_site_level_benchmarking_dataset
+Benchmarking variant set from the 1000 Genomes Project Phase 3 SV call set, see [Sudmant et al. 2015](https://doi.org/10.1038/nature15394).
+
+#### asc_site_level_benchmarking_dataset
+Benchmarking variant set from the Autism Spectrum Consortium. **Not public data.**
+
+#### hgsv_site_level_benchmarking_dataset
+Benchmarking variant set from [Werling et al. 2018](https://doi.org/10.1038%2Fs41588-018-0107-y). **Not public data.**
+
+#### collins_2017_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Collins et al. 2017](https://doi.org/10.1186/s13059-017-1158-6). **Not public data.**
+
+#### sanders_2015_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Sanders et al. 2015](https://doi.org/10.1016/j.neuron.2015.09.016). **Not public data.**
+
+#### werling_2018_sample_level_benchmarking_dataset
+Benchmarking genotypes from [Werling et al. 2018](https://doi.org/10.1038%2Fs41588-018-0107-y). **Not public data.**
+
diff --git a/website/docs/run/joint.md b/website/docs/run/joint.md
deleted file mode 100644
index 2d73959a3..000000000
--- a/website/docs/run/joint.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-title: Joint-calling
-description: Run the pipeline on a cohort
-sidebar_position: 4
-slug: joint
----
-
-:::info
-This documentation page is incomplete, and we are actively working on improving it with comprehensive information.
-:::
-
-## Batching
-
-For larger cohorts, samples should be split up into batches of about 100-500
-samples with similar characteristics. We recommend batching based on overall
-coverage and dosage score (WGD), which can be generated in [EvidenceQC](/docs/modules/eqc).
-An example batching process is outlined below:
-
-1. Divide the cohort into PCR+ and PCR- samples
-2. Partition the samples by median coverage from [EvidenceQC](/docs/modules/eqc),
- grouping samples with similar median coverage together. The end goal is to
- divide the cohort into roughly equal-sized batches of about 100-500 samples;
- if your partitions based on coverage are larger or uneven, you can partition
- the cohort further in the next step to obtain the final batches.
-3. Optionally, divide the samples further by dosage score (WGD) from
- [EvidenceQC](/docs/modules/eqc), grouping samples with similar WGD score
- together, to obtain roughly equal-sized batches of about 100-500 samples
-4. Maintain a roughly equal sex balance within each batch, based on sex
- assignments from [EvidenceQC](/docs/modules/eqc)
diff --git a/website/docs/run/overview.md b/website/docs/run/overview.md
deleted file mode 100644
index f9a82bcd1..000000000
--- a/website/docs/run/overview.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-title: Overview
-description: Overview
-sidebar_position: 1
-slug: overview
----
-
-There are two factors to consider when deciding how to run GATK-SV.
-
-
-1. **Variant calling modes: single-sample and cohort-based calling.**
- GATK-SV offers two distinct pipeline configurations for detecting
- structural variations (SVs), each tailored for different research needs:
-
- - **Single-sample analysis:**
- This configuration is ideal for examining SVs in individual samples,
- focusing exclusively on data from that single sample. Running this mode is less complex,
- involving just one workflow per sample.
-
- - **Joint calling:**
- This configuration is designed for more extensive studies, such as those
- involving population genetics or disease association studies.
- It analyzes SVs across a cohort by collectively assessing data from all samples.
- However, this comes with increased complexity compared to the single-sample mode,
- requiring the execution of multiple workflows and involves data preparation steps
- (e.g., batching files from the cohort).
-
-
-2. **Which platform you would like to use for running GATK-SV?**
- You may run GATK-SV on the following platforms.
-
- - [Terra.bio](https://terra.bio): A user-friendly cloud-native platform for scalable data analysis.
- The primary focus of this documentation is on supporting the execution of GATK-SV within the Terra platform.
-
- - [Cromwell](https://github.com/broadinstitute/cromwell):
- You may run GATK-SV on a self-hosted and managed cromwell instance, which is ideal for
- power-users and developers. We provide guidelines for this option in the
- [_advanced guides_](/docs/advanced/development/cromwell) section.
-
-
-Your decision regarding the execution modes and platform should be guided by
-the objectives of your study, the size of your cohort, data access needs,
-and the trade-off between a straightforward interface (Terra)
-and more detailed customization options (self-managed Cromwell server).
-Please refer to the following documentation on running GATK-SV within the Terra platform.
-
-- [Single-sample on Terra](single.md);
-- [Joint calling on Terra](joint.md).
diff --git a/website/docs/run/single.md b/website/docs/run/single.md
deleted file mode 100644
index 3f444b9d7..000000000
--- a/website/docs/run/single.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-title: Single-sample
-description: Run the pipeline on a single sample
-sidebar_position: 3
-slug: single
----
-
-:::info
-This documentation page is incomplete, and we are actively working on improving it with comprehensive information.
-:::
-
-We have developed a
- [Terra workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
-for running GATK-SV on a single sample, which contains the related documentation.
diff --git a/website/docs/runtime_attr.md b/website/docs/runtime_attr.md
new file mode 100644
index 000000000..be3cb6de2
--- /dev/null
+++ b/website/docs/runtime_attr.md
@@ -0,0 +1,51 @@
+---
+title: Runtime attributes
+description: Runtime attributes
+sidebar_position: 6
+---
+
+GATK-SV is implemented as a set of WDLs designed to run on Google Cloud Platform. Computations are broken up into
+a set of tasks that are carried out in a particular order on cloud virtual machines (VMs), each of which
+possesses a limited set of resources. These include the following primary components:
+
+- CPU cores
+- Random access memory (RAM)
+- Disk storage
+
+GATK-SV attempts to request the appropriate amount of resources for each task and in some cases employs mathematical
+models to predict optimal requirements that minimize cost. However, if the actual computations performed require more
+than the requested resources on the VM, the task may fail.
+
+:::info
+Most tasks in GATK-SV are tuned for cohorts with ~150 samples. Most tasks should scale automatically, but running larger
+cohorts may lead to slowdowns or errors due to insufficient resource allocation.
+:::
+
+In addition to VM resources, Terra and Cromwell both support automatic retries of tasks in case of ephemeral errors,
+as well as requesting [preemptible VMs](https://cloud.google.com/compute/docs/instances/preemptible) at a significant discount.
+
+### Setting runtime attributes
+
+GATK-SV exposes optional parameters for manually tuning VM resource allocation, automatic retries, and the use of preemptible
+instances. These parameters are all a custom struct called `RuntimeAttr`, which is defined in
+[Structs.wdl](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/Structs.wdl) as:
+
+```
+struct RuntimeAttr {
+ Float? mem_gb
+ Int? cpu_cores
+ Int? disk_gb
+ Int? boot_disk_gb
+ Int? preemptible_tries
+ Int? max_retries
+}
+```
+
+Users encountering errors due to insufficient VM resources or wishing to adjust automatic and preemptible retries may
+modify the corresponding parameters. Users should inspect the WDL source to determine whether a given `RuntimeAttr`
+parameter corresponds to a specific task.
+
+### Further reading
+
+For more information on identifying errors and setting runtime parameters, please refer to the
+[Troubleshooting FAQ](/docs/troubleshooting/faq).
diff --git a/website/docs/troubleshooting/_category_.json b/website/docs/troubleshooting/_category_.json
index 9bcfbef40..09047afec 100644
--- a/website/docs/troubleshooting/_category_.json
+++ b/website/docs/troubleshooting/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Troubleshooting",
- "position": 5,
+ "position": 8,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/troubleshooting/faq.md b/website/docs/troubleshooting/faq.md
index 1cd431028..439cbf391 100644
--- a/website/docs/troubleshooting/faq.md
+++ b/website/docs/troubleshooting/faq.md
@@ -3,12 +3,17 @@ title: FAQ
slug: faq
---
+Please consult the following resources for additional troubleshooting guides:
+
+- [Troubleshooting GATK-SV (article)](https://gatk.broadinstitute.org/hc/en-us/articles/5334566940699-Troubleshooting-GATK-SV)
+- [Troubleshooting GATK-SV Error Messages on Terra (video)](https://www.youtube.com/watch?v=3UVV03H9p1w)
+
### VM runs out of memory or disk
-- Default pipeline settings are tuned for batches of 100 samples.
+- Default pipeline settings are tuned for batches of 150 samples.
Larger batches or cohorts may require additional VM resources.
Most runtime attributes can be modified through
- the RuntimeAttr inputs. These are formatted like this in the json:
+ the `RuntimeAttr` inputs. These can be formatted like this in an input json:
```json
"MyWorkflow.runtime_attr_override": {
@@ -18,7 +23,7 @@ slug: faq
```
Note that a subset of the struct attributes can be specified.
- See `wdl/Structs.wdl` for available attributes.
+ See [wdl/Structs.wdl](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/Structs.wdl) for available attributes.
### Calculated read length causes error in MELT workflow
diff --git a/website/package.json b/website/package.json
index fa32a8117..70fea28a2 100644
--- a/website/package.json
+++ b/website/package.json
@@ -14,9 +14,9 @@
"write-heading-ids": "docusaurus write-heading-ids"
},
"dependencies": {
- "@docusaurus/core": "3.3.2",
- "@docusaurus/preset-classic": "3.3.2",
- "@docusaurus/theme-mermaid": "3.3.2",
+ "@docusaurus/core": "^3.5.2",
+ "@docusaurus/preset-classic": "^3.5.2",
+ "@docusaurus/theme-mermaid": "^3.5.2",
"@mdx-js/react": "^3.0.0",
"clsx": "^2.0.0",
"prism-react-renderer": "^2.3.0",
@@ -24,8 +24,8 @@
"react-dom": "^18.0.0"
},
"devDependencies": {
- "@docusaurus/module-type-aliases": "3.3.2",
- "@docusaurus/types": "3.3.2"
+ "@docusaurus/module-type-aliases": "^3.5.2",
+ "@docusaurus/types": "^3.5.2"
},
"browserslist": {
"production": [
diff --git a/website/static/img/qc/VcfQcGenotypeDistributions.png b/website/static/img/qc/VcfQcGenotypeDistributions.png
new file mode 100644
index 000000000..c18ae574d
--- /dev/null
+++ b/website/static/img/qc/VcfQcGenotypeDistributions.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:23e7e565ed6550ea79332bef7dea04a1fff2ed28932b96d694d8de5d7e690352
+size 1115174
diff --git a/website/static/img/qc/VcfQcGnomADv2CollinsSVAllCallsetBenchmarking.png b/website/static/img/qc/VcfQcGnomADv2CollinsSVAllCallsetBenchmarking.png
new file mode 100644
index 000000000..d89e0d16a
--- /dev/null
+++ b/website/static/img/qc/VcfQcGnomADv2CollinsSVAllCallsetBenchmarking.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:9cc3dfca00798620a6b2b90bbd51fd0b741800b9b4c768556b4d7aac763a1c9d
+size 1064628
diff --git a/website/static/img/qc/VcfQcSizeDistributionsMerged.png b/website/static/img/qc/VcfQcSizeDistributionsMerged.png
new file mode 100644
index 000000000..4d468cdf3
--- /dev/null
+++ b/website/static/img/qc/VcfQcSizeDistributionsMerged.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:561a681de80ee93f8a1dfc47b0086828dde6c121223c4ebff6d47a73e88697e8
+size 151637
diff --git a/website/static/img/qc/VcfQcSvCountsMerged.png b/website/static/img/qc/VcfQcSvCountsMerged.png
new file mode 100644
index 000000000..7d2c5fd77
--- /dev/null
+++ b/website/static/img/qc/VcfQcSvCountsMerged.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:64a882fe1ca04ec7da830fb249c2799e5fbab052951a476facfe2eca01c5214b
+size 40665
diff --git a/website/static/img/qc/VcfQcSvPerGenome.png b/website/static/img/qc/VcfQcSvPerGenome.png
new file mode 100644
index 000000000..15d06f19c
--- /dev/null
+++ b/website/static/img/qc/VcfQcSvPerGenome.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:b04d0ac119d7738f5c084db546df86d6c55d030ca72eea10807f390af4b87fe5
+size 702941
diff --git a/website/static/img/qc/VcfQcSvTrioInheritance.png b/website/static/img/qc/VcfQcSvTrioInheritance.png
new file mode 100644
index 000000000..44cc1ad59
--- /dev/null
+++ b/website/static/img/qc/VcfQcSvTrioInheritance.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:068adff3b217398fe4f410cdc70ac128a56bce44ea811f58114c518c9547a692
+size 134280
diff --git a/website/static/img/qc/VcfQqFreqDistributionsMerged.png b/website/static/img/qc/VcfQqFreqDistributionsMerged.png
new file mode 100644
index 000000000..7661d2eb2
--- /dev/null
+++ b/website/static/img/qc/VcfQqFreqDistributionsMerged.png
@@ -0,0 +1,3 @@
+version https://git-lfs.github.com/spec/v1
+oid sha256:6618befbf0a05ebd683783995d974e0bb5720f3f41c39268c1c18f3b091154eb
+size 97067
 +
+The following workflows are included in this workspace, to be executed in this order:
+
+1. `01-GatherSampleEvidence`: Per-sample SV evidence collection, including calls from a configurable set of
+algorithms (Manta, MELT, and Wham), read depth (RD), split read positions (SR), and discordant pair positions (PE).
+2. `02-EvidenceQC`: Dosage bias scoring and ploidy estimation, run on preliminary batches
+3. `03-TrainGCNV`: Per-batch training of a gCNV model for use in `04-GatherBatchEvidence`
+4. `04-GatherBatchEvidence`: Per-batch copy number variant calling using cn.MOPS and GATK gCNV; B-allele frequency (BAF)
+generation; call and evidence aggregation
+5. `05-ClusterBatch`: Per-batch variant clustering
+6. `06-GenerateBatchMetrics`: Per-batch variant filtering, metric generation
+7. `07-FilterBatchSites`: Per-batch variant filtering and plot SV counts per sample per SV type to enable choice of IQR
+cutoff for outlier filtration in `08-FilterBatchSamples`
+8. `08-FilterBatchSamples`: Per-batch outlier sample filtration
+9. `09-MergeBatchSites`: Site merging of SVs discovered across batches, run on a cohort-level `sample_set_set`
+10. `10-GenotypeBatch`: Per-batch genotyping of all sites in the cohort
+11. `11-RegenotypeCNVs`: Cohort-level genotype refinement of some depth calls
+12. `12-CombineBatches`: Cohort-level cross-batch integration and clustering
+13. `13-ResolveComplexVariants`: Complex variant resolution
+14. `14-GenotypeComplexVariants`: Complex variant re-genotyping
+15. `15-CleanVcf`: VCF cleanup
+16. `16-RefineComplexVariants`: Complex variant filtering and refinement
+17. `17-ApplyManualVariantFilter`: Hard filtering high-FP SV classes
+18. `18-JoinRawCalls`: Raw call aggregation
+19. `19-SVConcordance`: Annotate genotype concordance with raw calls
+20. `20-FilterGenotypes`: Genotype filtering
+21. `21-AnnotateVcf`: Cohort VCF annotations, including functional annotation, allele frequency (AF) annotation, and
+AF annotation with external population callsets
+
+Extra workflows (Not part of canonical pipeline, but included for your convenience. May require manual configuration):
+* `MainVcfQc`: Generate detailed call set QC plots
+* `PlotSVCountsPerSample`: Plot SV counts per sample per SV type. Recommended to run before `FilterOutlierSamples`
+ (configured with the single VCF you want to filter) to enable IQR cutoff choice.
+* `FilterOutlierSamples`: Filter outlier samples (in terms of SV counts) from a single VCF.
+* `VisualizeCnvs`: Plot multi-sample depth profiles for CNVs
+
+For detailed instructions on running the pipeline in Terra, see [workflow instructions](#instructions) below.
+
+#### What is the maximum number of samples the pipeline can handle?
+
+In Terra, we have tested batch sizes of up to 500 samples and cohort sizes (consisting of multiple batches) of up to
+11,000 samples (and 98,000 samples with the final steps split by chromosome). On a dedicated Cromwell server, we have
+tested the pipeline on cohorts of up to ~140,000 samples.
+
+
+### Time and cost estimates
+
+The following estimates pertain to the 1000 Genomes sample data in this workspace. They represent aggregated run time
+and cost across modules for the whole pipeline. For workflows run multiple times (on each sample or on each batch),
+the longest individual runtime was used. Call caching may affect some of this information.
+
+|Number of samples|Time|Total run cost|Per-sample run cost|
+|--------------|--------|----------|----------|
+|312|~76 hours|~$675|~$2.16/sample|
+
+Please note that sample characteristics, cohort size, and level of filtering may influence pipeline compute costs,
+with average costs ranging between $2-$3 per sample. For instance, PCR+ samples and samples with a high percentage
+of improperly paired reads have been observed to cost more. Consider
+[excluding low-quality samples](/docs/gs/inputs#sample-exclusion) prior to processing to keep costs low.
+
+### Sample ID format
+
+Refer to [the sample ID requirements section](/docs/gs/inputs#sampleids) of the documentation.
+
+### Workspace setup
+
+1. Clone this workspace into a Terra project to which you have access
+
+2. In your new workspace, delete the example data. To do this, go to the *Data* tab of the workspace. Delete the data
+ tables in this order: `sample_set_set`, `sample_set`, and `sample`. For each table, click the 3 dots icon to the
+ right of the table name and click "Delete table". Confirm when prompted.
+
+
+The following workflows are included in this workspace, to be executed in this order:
+
+1. `01-GatherSampleEvidence`: Per-sample SV evidence collection, including calls from a configurable set of
+algorithms (Manta, MELT, and Wham), read depth (RD), split read positions (SR), and discordant pair positions (PE).
+2. `02-EvidenceQC`: Dosage bias scoring and ploidy estimation, run on preliminary batches
+3. `03-TrainGCNV`: Per-batch training of a gCNV model for use in `04-GatherBatchEvidence`
+4. `04-GatherBatchEvidence`: Per-batch copy number variant calling using cn.MOPS and GATK gCNV; B-allele frequency (BAF)
+generation; call and evidence aggregation
+5. `05-ClusterBatch`: Per-batch variant clustering
+6. `06-GenerateBatchMetrics`: Per-batch variant filtering, metric generation
+7. `07-FilterBatchSites`: Per-batch variant filtering and plot SV counts per sample per SV type to enable choice of IQR
+cutoff for outlier filtration in `08-FilterBatchSamples`
+8. `08-FilterBatchSamples`: Per-batch outlier sample filtration
+9. `09-MergeBatchSites`: Site merging of SVs discovered across batches, run on a cohort-level `sample_set_set`
+10. `10-GenotypeBatch`: Per-batch genotyping of all sites in the cohort
+11. `11-RegenotypeCNVs`: Cohort-level genotype refinement of some depth calls
+12. `12-CombineBatches`: Cohort-level cross-batch integration and clustering
+13. `13-ResolveComplexVariants`: Complex variant resolution
+14. `14-GenotypeComplexVariants`: Complex variant re-genotyping
+15. `15-CleanVcf`: VCF cleanup
+16. `16-RefineComplexVariants`: Complex variant filtering and refinement
+17. `17-ApplyManualVariantFilter`: Hard filtering high-FP SV classes
+18. `18-JoinRawCalls`: Raw call aggregation
+19. `19-SVConcordance`: Annotate genotype concordance with raw calls
+20. `20-FilterGenotypes`: Genotype filtering
+21. `21-AnnotateVcf`: Cohort VCF annotations, including functional annotation, allele frequency (AF) annotation, and
+AF annotation with external population callsets
+
+Extra workflows (Not part of canonical pipeline, but included for your convenience. May require manual configuration):
+* `MainVcfQc`: Generate detailed call set QC plots
+* `PlotSVCountsPerSample`: Plot SV counts per sample per SV type. Recommended to run before `FilterOutlierSamples`
+ (configured with the single VCF you want to filter) to enable IQR cutoff choice.
+* `FilterOutlierSamples`: Filter outlier samples (in terms of SV counts) from a single VCF.
+* `VisualizeCnvs`: Plot multi-sample depth profiles for CNVs
+
+For detailed instructions on running the pipeline in Terra, see [workflow instructions](#instructions) below.
+
+#### What is the maximum number of samples the pipeline can handle?
+
+In Terra, we have tested batch sizes of up to 500 samples and cohort sizes (consisting of multiple batches) of up to
+11,000 samples (and 98,000 samples with the final steps split by chromosome). On a dedicated Cromwell server, we have
+tested the pipeline on cohorts of up to ~140,000 samples.
+
+
+### Time and cost estimates
+
+The following estimates pertain to the 1000 Genomes sample data in this workspace. They represent aggregated run time
+and cost across modules for the whole pipeline. For workflows run multiple times (on each sample or on each batch),
+the longest individual runtime was used. Call caching may affect some of this information.
+
+|Number of samples|Time|Total run cost|Per-sample run cost|
+|--------------|--------|----------|----------|
+|312|~76 hours|~$675|~$2.16/sample|
+
+Please note that sample characteristics, cohort size, and level of filtering may influence pipeline compute costs,
+with average costs ranging between $2-$3 per sample. For instance, PCR+ samples and samples with a high percentage
+of improperly paired reads have been observed to cost more. Consider
+[excluding low-quality samples](/docs/gs/inputs#sample-exclusion) prior to processing to keep costs low.
+
+### Sample ID format
+
+Refer to [the sample ID requirements section](/docs/gs/inputs#sampleids) of the documentation.
+
+### Workspace setup
+
+1. Clone this workspace into a Terra project to which you have access
+
+2. In your new workspace, delete the example data. To do this, go to the *Data* tab of the workspace. Delete the data
+ tables in this order: `sample_set_set`, `sample_set`, and `sample`. For each table, click the 3 dots icon to the
+ right of the table name and click "Delete table". Confirm when prompted.
+  +
+3. Create and upload a new sample data table for your samples. This should be a tab-separated file (.tsv) with one line
+ per sample, as well as a header (first) line. It should contain the columns `entity:sample_id` (first column) and
+ `bam_or_cram_file` at minimum. See the **Required inputs** section above for more information on these inputs. For
+ an example sample data table, refer to the sample data table for the 1000 Genomes samples in this workspace
+ [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/samples_1kgp.tsv.tmpl).
+ To upload the TSV file, navigate to the *Data* tab of the workspace, click the `Import Data` button on the top left,
+ and select "Upload TSV".
+
+
+3. Create and upload a new sample data table for your samples. This should be a tab-separated file (.tsv) with one line
+ per sample, as well as a header (first) line. It should contain the columns `entity:sample_id` (first column) and
+ `bam_or_cram_file` at minimum. See the **Required inputs** section above for more information on these inputs. For
+ an example sample data table, refer to the sample data table for the 1000 Genomes samples in this workspace
+ [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/samples_1kgp.tsv.tmpl).
+ To upload the TSV file, navigate to the *Data* tab of the workspace, click the `Import Data` button on the top left,
+ and select "Upload TSV".
+  +
+4. Edit the `cohort_ped_file` item in the Workspace Data table (as shown in the screenshot below) to provide the Google
+ URI to the PED file for your cohort (make sure to share it with your Terra proxy account!).
+
+
+4. Edit the `cohort_ped_file` item in the Workspace Data table (as shown in the screenshot below) to provide the Google
+ URI to the PED file for your cohort (make sure to share it with your Terra proxy account!).
+  +
+
+### Creating sample_sets
+
+To create batches (in the `sample_set` table), the easiest way is to upload a tab-separated sample set membership file.
+This file should have one line per sample, plus a header (first) line. The first column should be
+`membership:sample_set_id` (containing the `sample_set_id` for the sample in question), and the second should be
+`sample` (containing the sample IDs). Recall that batch IDs (`sample_set_id`) should follow the
+[sample ID requirements](/docs/gs/inputs#sampleids). For an example sample membership file, refer to the one for the
+1000 Genomes samples in this workspace [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/sample_set_membership_1kgp.tsv.tmpl).
+
+## Workflow instructions {#instructions}
+
+### General recommendations
+
+* It is recommended to run each workflow first on one sample/batch to check that the method is properly configured
+before you attempt to process all of your data.
+* We recommend enabling call-caching (on by default in each workflow configuration).
+* We recommend enabling automatic intermediate file deletion by checking the box labeled "Delete intermediate outputs"
+at the top of the workflow launch page every time you start a workflow. With this option enabled, intermediate files
+(those not present in the Terra data table, and not needed for any further GATK-SV processing) will be deleted
+automatically if the workflow succeeds. If the workflow fails, the outputs will be retained to enable a re-run to
+pick up where it left off with call-caching. However, call-caching will not be possible for workflows that have
+succeeded. For more information on this option, see
+[this article](https://terra.bio/delete-intermediates-option-now-available-for-workflows-in-terra/). For guidance on
+managing intermediate storage from failed workflows, or from workflows without the delete intermediates option enabled,
+see the next bullet point.
+* There are cases when you may need to manage storage in other ways: for workflows that failed (only delete files from
+a failed workflow after a version has succeeded, to avoid disabling call-caching), for workflows without intermediate
+file deletion enabled, or once you are done processing and want to delete files from earlier steps in the pipeline
+that you no longer need.
+ * One option is to manually delete large files, or directories containing failed workflow intermediates (after
+ re-running the workflow successfully to take advantage of call-caching) with the command
+ `gsutil -m rm gs://path/to/workflow/directory/**file_extension_to_delete` to delete all files with the given extension
+ for that workflow, or `gsutil -m rm -r gs://path/to/workflow/directory/` to delete an entire workflow directory
+ (only after you are done with all the files!). Note that this can take a very long time for larger workflows, which
+ may contain thousands of files.
+ * Another option is to use the `fiss mop` API call to delete all files that do not appear in one of the Terra data
+ tables (intermediate files). Always ensure that you are completely done with a step and you will not need to return
+ before using this option, as it will break call-caching. See
+ [this blog post](https://terra.bio/deleting-intermediate-workflow-outputs/) for more details. This can also be done
+ [via the command line](https://github.com/broadinstitute/fiss/wiki/MOP:-reducing-your-cloud-storage-footprint).
+* If your workflow fails, check the job manager for the error message. Most issues can be resolved by increasing the
+memory or disk. Do not delete workflow log files until you are done troubleshooting. If call-caching is enabled, do not
+delete any files from the failed workflow until you have run it successfully.
+* To display run costs, see [this article](https://support.terra.bio/hc/en-us/articles/360037862771#h_01EX5ED53HAZ59M29DRCG24CXY)
+for one-time setup instructions for non-Broad users.
+
+### 01-GatherSampleEvidence
+
+Read the full GatherSampleEvidence documentation [here](/docs/modules/gse).
+* This workflow runs on a per-sample level, but you can launch many (a few hundred) samples at once, in arbitrary
+partitions. Make sure to try just one sample first though!
+* Refer to the [Input Data section](/docs/gs/inputs) for details on file formats, sample QC, and sample ID restrictions.
+* It is normal for a few samples in a cohort to run out of memory during Wham SV calling, so we recommend enabling
+auto-retry for out-of-memory errors for `01-GatherSampleEvidence` only. Before you launch the workflow, click the
+checkbox reading "Retry with more memory" and set the memory retry factor to 1.8. This action must be performed each
+time you launch a `01-GatherSampleEvidence` job.
+* Please note that most large published joint call sets produced by GATK-SV, including gnomAD-SV, included the tool
+MELT, a state-of-the-art mobile element insertion (MEI) detector, as part of the pipeline. Due to licensing
+restrictions, we cannot provide a public docker image for this algorithm. The `01-GatherSampleEvidence` workflow
+does not use MELT as one of the SV callers by default, which will result in less sensitivity to MEI calls. In order
+to use MELT, you will need to build your own private docker image (example Dockerfile
+[here](https://github.com/broadinstitute/gatk-sv/blob/master/dockerfiles/melt/Dockerfile)), share it with your Terra
+proxy account, enter it in the `melt_docker` input in the `01-GatherSampleEvidence` configuration (as a string,
+surrounded by double-quotes), and then click "Save". No further changes are necessary beyond `01-GatherSampleEvidence`.
+ * Note that the version of MELT tested with GATK-SV is v2.0.5. If you use a different version to create your own
+ docker image, we recommend testing your image by running one pilot sample through `01-GatherSampleEvidence` to check
+ that it runs as expected, then running a small group of about 10 pilot samples through the pipeline until the end of
+ `04-GatherBatchEvidence` to check that the outputs are compatible with GATK-SV.
+* If you enable "Delete intermediate outputs" whenever you launch this workflow (recommended), BAM files will be
+deleted for successful runs; but BAM files will not be deleted if the run fails or if intermediate file deletion is
+not enabled. Since BAM files are large, we recommend deleting them to save on storage costs, but only after fixing and
+re-running the failed workflow, so that it will call-cache.
+
+
+### 02-EvidenceQC
+
+Read the full EvidenceQC documentation [here](/docs/modules/eqc).
+* `02-EvidenceQC` is run on arbitrary cohort partitions of up to 500 samples.
+* The outputs from `02-EvidenceQC` can be used for
+[preliminary sample QC](/docs/modules/eqc#preliminary-sample-qc) and
+[batching](#batching) before moving on to [TrainGCNV](#traingcnv).
+
+
+### Batching (manual step) {#batching}
+
+For larger cohorts, samples should be split up into batches of about 100-500
+samples with similar characteristics. We recommend batching based on overall
+coverage and dosage score (WGD), which can be generated in [EvidenceQC](/docs/modules/eqc).
+An example batching process is outlined below:
+
+1. Divide the cohort into PCR+ and PCR- samples
+2. Partition the samples by median coverage from [EvidenceQC](/docs/modules/eqc),
+ grouping samples with similar median coverage together. The end goal is to
+ divide the cohort into roughly equal-sized batches of about 100-500 samples;
+ if your partitions based on coverage are larger or uneven, you can partition
+ the cohort further in the next step to obtain the final batches.
+3. Optionally, divide the samples further by dosage score (WGD) from
+ [EvidenceQC](/docs/modules/eqc), grouping samples with similar WGD score
+ together, to obtain roughly equal-sized batches of about 100-500 samples
+4. Maintain a roughly equal sex balance within each batch, based on sex
+ assignments from [EvidenceQC](/docs/modules/eqc)
+
+
+### 03-TrainGCNV {#traingcnv}
+
+Read the full TrainGCNV documentation [here](/docs/modules/gcnv).
+* Before running this workflow, create the batches (~100-500 samples) you will use for the rest of the pipeline based
+on sample coverage, WGD score (from `02-EvidenceQC`), and PCR status. These will likely not be the same as the batches
+you used for `02-EvidenceQC`.
+* By default, `03-TrainGCNV` is configured to be run once per `sample_set` on 100 randomly-chosen samples from that
+set to create a gCNV model for each batch. To modify this behavior, you can set the `n_samples_subsample` parameter
+to the number of samples to use for training.
+
+### 04-GatherBatchEvidence
+
+Read the full GatherBatchEvidence documentation [here](/docs/modules/gbe).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV`.
+* Before running this workflow, ensure that you have updated the `cohort_ped_file` attribute in Workspace Data with
+your cohort's PED file, with sex assignments updated based on ploidy detection from `02-EvidenceQC`.
+
+### Steps 05-06
+
+Read the full documentation for [ClusterBatch](/docs/modules/cb) and [GenerateBatchMetrics](/docs/modules/gbm).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` and `04-GatherBatchEvidence`.
+
+
+### Steps 07-08
+
+These two workflows make up FilterBatch; they are subdivided in this workspace to enable tuning of outlier filtration
+cutoffs. Read the full FilterBatch documentation [here](/docs/modules/fb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `06-GenerateBatchMetrics`.
+* `07-FilterBatchSites` produces SV count plots and files, as well as a preview of the outlier samples to be filtered.
+The input `N_IQR_cutoff_plotting` is used to visualize filtration thresholds on the SV count plots and preview the
+samples to be filtered; the default value is set to 6. You can adjust this value depending on your needs, and you can
+re-run the workflow with new `N_IQR_cutoff_plotting` values until the plots and outlier sample lists suit the purposes
+of your study. Once you have chosen an IQR cutoff, provide it to the `N_IQR_cutoff` input in `08-FilterBatchSamples` to
+filter the VCFs using the chosen cutoff.
+* `08-FilterBatchSamples` performs outlier sample filtration, removing samples with an abnormal number of SV calls of
+at least one SV type. To tune the filtering threshold to your needs, edit the `N_IQR_cutoff` input value based on the
+plots and outlier sample preview lists from `07-FilterBatchSites`. The default value for `N_IQR_cutoff` in this step
+is 10000, which essentially means that no samples are filtered.
+
+### 09-MergeBatchSites
+
+Read the full MergeBatchSites documentation [here](/docs/modules/msites).
+* `09-MergeBatchSites` is a cohort-level workflow, so it is run on a `sample_set_set` containing all the batches
+in the cohort. Navigate to the Data tab of your workspace. If there is no `sample_set_set` data table, you will need
+to create it. To do this, select the `sample_set` data table, then select (with the checkboxes) all the batches
+(`sample_set`) in your cohort. These should be the `sample_sets` that you used to run steps `03-TrainGCNV` through
+`08-FilterBatchSamples`. Then click the "Edit" icon above the table and choose "Save selection as set." Enter a name
+that follows the **Sample ID requirements**. This will create a new `sample_set_set` containing all of the `sample_sets`
+in your cohort. When you launch MergeBatchSites, you can now select this `sample_set_set`.
+
+
+
+
+### Creating sample_sets
+
+To create batches (in the `sample_set` table), the easiest way is to upload a tab-separated sample set membership file.
+This file should have one line per sample, plus a header (first) line. The first column should be
+`membership:sample_set_id` (containing the `sample_set_id` for the sample in question), and the second should be
+`sample` (containing the sample IDs). Recall that batch IDs (`sample_set_id`) should follow the
+[sample ID requirements](/docs/gs/inputs#sampleids). For an example sample membership file, refer to the one for the
+1000 Genomes samples in this workspace [here in the GATK-SV GitHub repository](https://github.com/broadinstitute/gatk-sv/blob/master/input_templates/terra_workspaces/cohort_mode/sample_set_membership_1kgp.tsv.tmpl).
+
+## Workflow instructions {#instructions}
+
+### General recommendations
+
+* It is recommended to run each workflow first on one sample/batch to check that the method is properly configured
+before you attempt to process all of your data.
+* We recommend enabling call-caching (on by default in each workflow configuration).
+* We recommend enabling automatic intermediate file deletion by checking the box labeled "Delete intermediate outputs"
+at the top of the workflow launch page every time you start a workflow. With this option enabled, intermediate files
+(those not present in the Terra data table, and not needed for any further GATK-SV processing) will be deleted
+automatically if the workflow succeeds. If the workflow fails, the outputs will be retained to enable a re-run to
+pick up where it left off with call-caching. However, call-caching will not be possible for workflows that have
+succeeded. For more information on this option, see
+[this article](https://terra.bio/delete-intermediates-option-now-available-for-workflows-in-terra/). For guidance on
+managing intermediate storage from failed workflows, or from workflows without the delete intermediates option enabled,
+see the next bullet point.
+* There are cases when you may need to manage storage in other ways: for workflows that failed (only delete files from
+a failed workflow after a version has succeeded, to avoid disabling call-caching), for workflows without intermediate
+file deletion enabled, or once you are done processing and want to delete files from earlier steps in the pipeline
+that you no longer need.
+ * One option is to manually delete large files, or directories containing failed workflow intermediates (after
+ re-running the workflow successfully to take advantage of call-caching) with the command
+ `gsutil -m rm gs://path/to/workflow/directory/**file_extension_to_delete` to delete all files with the given extension
+ for that workflow, or `gsutil -m rm -r gs://path/to/workflow/directory/` to delete an entire workflow directory
+ (only after you are done with all the files!). Note that this can take a very long time for larger workflows, which
+ may contain thousands of files.
+ * Another option is to use the `fiss mop` API call to delete all files that do not appear in one of the Terra data
+ tables (intermediate files). Always ensure that you are completely done with a step and you will not need to return
+ before using this option, as it will break call-caching. See
+ [this blog post](https://terra.bio/deleting-intermediate-workflow-outputs/) for more details. This can also be done
+ [via the command line](https://github.com/broadinstitute/fiss/wiki/MOP:-reducing-your-cloud-storage-footprint).
+* If your workflow fails, check the job manager for the error message. Most issues can be resolved by increasing the
+memory or disk. Do not delete workflow log files until you are done troubleshooting. If call-caching is enabled, do not
+delete any files from the failed workflow until you have run it successfully.
+* To display run costs, see [this article](https://support.terra.bio/hc/en-us/articles/360037862771#h_01EX5ED53HAZ59M29DRCG24CXY)
+for one-time setup instructions for non-Broad users.
+
+### 01-GatherSampleEvidence
+
+Read the full GatherSampleEvidence documentation [here](/docs/modules/gse).
+* This workflow runs on a per-sample level, but you can launch many (a few hundred) samples at once, in arbitrary
+partitions. Make sure to try just one sample first though!
+* Refer to the [Input Data section](/docs/gs/inputs) for details on file formats, sample QC, and sample ID restrictions.
+* It is normal for a few samples in a cohort to run out of memory during Wham SV calling, so we recommend enabling
+auto-retry for out-of-memory errors for `01-GatherSampleEvidence` only. Before you launch the workflow, click the
+checkbox reading "Retry with more memory" and set the memory retry factor to 1.8. This action must be performed each
+time you launch a `01-GatherSampleEvidence` job.
+* Please note that most large published joint call sets produced by GATK-SV, including gnomAD-SV, included the tool
+MELT, a state-of-the-art mobile element insertion (MEI) detector, as part of the pipeline. Due to licensing
+restrictions, we cannot provide a public docker image for this algorithm. The `01-GatherSampleEvidence` workflow
+does not use MELT as one of the SV callers by default, which will result in less sensitivity to MEI calls. In order
+to use MELT, you will need to build your own private docker image (example Dockerfile
+[here](https://github.com/broadinstitute/gatk-sv/blob/master/dockerfiles/melt/Dockerfile)), share it with your Terra
+proxy account, enter it in the `melt_docker` input in the `01-GatherSampleEvidence` configuration (as a string,
+surrounded by double-quotes), and then click "Save". No further changes are necessary beyond `01-GatherSampleEvidence`.
+ * Note that the version of MELT tested with GATK-SV is v2.0.5. If you use a different version to create your own
+ docker image, we recommend testing your image by running one pilot sample through `01-GatherSampleEvidence` to check
+ that it runs as expected, then running a small group of about 10 pilot samples through the pipeline until the end of
+ `04-GatherBatchEvidence` to check that the outputs are compatible with GATK-SV.
+* If you enable "Delete intermediate outputs" whenever you launch this workflow (recommended), BAM files will be
+deleted for successful runs; but BAM files will not be deleted if the run fails or if intermediate file deletion is
+not enabled. Since BAM files are large, we recommend deleting them to save on storage costs, but only after fixing and
+re-running the failed workflow, so that it will call-cache.
+
+
+### 02-EvidenceQC
+
+Read the full EvidenceQC documentation [here](/docs/modules/eqc).
+* `02-EvidenceQC` is run on arbitrary cohort partitions of up to 500 samples.
+* The outputs from `02-EvidenceQC` can be used for
+[preliminary sample QC](/docs/modules/eqc#preliminary-sample-qc) and
+[batching](#batching) before moving on to [TrainGCNV](#traingcnv).
+
+
+### Batching (manual step) {#batching}
+
+For larger cohorts, samples should be split up into batches of about 100-500
+samples with similar characteristics. We recommend batching based on overall
+coverage and dosage score (WGD), which can be generated in [EvidenceQC](/docs/modules/eqc).
+An example batching process is outlined below:
+
+1. Divide the cohort into PCR+ and PCR- samples
+2. Partition the samples by median coverage from [EvidenceQC](/docs/modules/eqc),
+ grouping samples with similar median coverage together. The end goal is to
+ divide the cohort into roughly equal-sized batches of about 100-500 samples;
+ if your partitions based on coverage are larger or uneven, you can partition
+ the cohort further in the next step to obtain the final batches.
+3. Optionally, divide the samples further by dosage score (WGD) from
+ [EvidenceQC](/docs/modules/eqc), grouping samples with similar WGD score
+ together, to obtain roughly equal-sized batches of about 100-500 samples
+4. Maintain a roughly equal sex balance within each batch, based on sex
+ assignments from [EvidenceQC](/docs/modules/eqc)
+
+
+### 03-TrainGCNV {#traingcnv}
+
+Read the full TrainGCNV documentation [here](/docs/modules/gcnv).
+* Before running this workflow, create the batches (~100-500 samples) you will use for the rest of the pipeline based
+on sample coverage, WGD score (from `02-EvidenceQC`), and PCR status. These will likely not be the same as the batches
+you used for `02-EvidenceQC`.
+* By default, `03-TrainGCNV` is configured to be run once per `sample_set` on 100 randomly-chosen samples from that
+set to create a gCNV model for each batch. To modify this behavior, you can set the `n_samples_subsample` parameter
+to the number of samples to use for training.
+
+### 04-GatherBatchEvidence
+
+Read the full GatherBatchEvidence documentation [here](/docs/modules/gbe).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV`.
+* Before running this workflow, ensure that you have updated the `cohort_ped_file` attribute in Workspace Data with
+your cohort's PED file, with sex assignments updated based on ploidy detection from `02-EvidenceQC`.
+
+### Steps 05-06
+
+Read the full documentation for [ClusterBatch](/docs/modules/cb) and [GenerateBatchMetrics](/docs/modules/gbm).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` and `04-GatherBatchEvidence`.
+
+
+### Steps 07-08
+
+These two workflows make up FilterBatch; they are subdivided in this workspace to enable tuning of outlier filtration
+cutoffs. Read the full FilterBatch documentation [here](/docs/modules/fb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `06-GenerateBatchMetrics`.
+* `07-FilterBatchSites` produces SV count plots and files, as well as a preview of the outlier samples to be filtered.
+The input `N_IQR_cutoff_plotting` is used to visualize filtration thresholds on the SV count plots and preview the
+samples to be filtered; the default value is set to 6. You can adjust this value depending on your needs, and you can
+re-run the workflow with new `N_IQR_cutoff_plotting` values until the plots and outlier sample lists suit the purposes
+of your study. Once you have chosen an IQR cutoff, provide it to the `N_IQR_cutoff` input in `08-FilterBatchSamples` to
+filter the VCFs using the chosen cutoff.
+* `08-FilterBatchSamples` performs outlier sample filtration, removing samples with an abnormal number of SV calls of
+at least one SV type. To tune the filtering threshold to your needs, edit the `N_IQR_cutoff` input value based on the
+plots and outlier sample preview lists from `07-FilterBatchSites`. The default value for `N_IQR_cutoff` in this step
+is 10000, which essentially means that no samples are filtered.
+
+### 09-MergeBatchSites
+
+Read the full MergeBatchSites documentation [here](/docs/modules/msites).
+* `09-MergeBatchSites` is a cohort-level workflow, so it is run on a `sample_set_set` containing all the batches
+in the cohort. Navigate to the Data tab of your workspace. If there is no `sample_set_set` data table, you will need
+to create it. To do this, select the `sample_set` data table, then select (with the checkboxes) all the batches
+(`sample_set`) in your cohort. These should be the `sample_sets` that you used to run steps `03-TrainGCNV` through
+`08-FilterBatchSamples`. Then click the "Edit" icon above the table and choose "Save selection as set." Enter a name
+that follows the **Sample ID requirements**. This will create a new `sample_set_set` containing all of the `sample_sets`
+in your cohort. When you launch MergeBatchSites, you can now select this `sample_set_set`.
+
+ +
+ +* If there is already a `sample_set_set` data table in your workspace, you can create this `sample_set_set` while you
+are launching the `09-MergeBatchSites` workflow: click "Select Data", choose "Create new sample_set_set [...]", check
+all the batches to include (all the ones used in `03-TrainGCNV` through `08-FilterBatchSamples`), and give it a name
+that follows the [sample ID requirements](/docs/gs/inputs#sampleids).
+
+
+* If there is already a `sample_set_set` data table in your workspace, you can create this `sample_set_set` while you
+are launching the `09-MergeBatchSites` workflow: click "Select Data", choose "Create new sample_set_set [...]", check
+all the batches to include (all the ones used in `03-TrainGCNV` through `08-FilterBatchSamples`), and give it a name
+that follows the [sample ID requirements](/docs/gs/inputs#sampleids).
+
+ +
+### 10-GenotypeBatch
+
+Read the full GenotypeBatch documentation [here](/docs/modules/gb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `08-FilterBatchSamples`.
+
+### Steps 11-17
+
+Read the full documentation for [RegenotypeCNVs](/docs/modules/rgcnvs), [MakeCohortVcf](/docs/modules/cvcf) (which
+includes `CombineBatches`, `ResolveComplexVariants`, `GenotypeComplexVariants`, `CleanVcf`, `MainVcfQc`), and
+[AnnotateVcf](/docs/modules/av).
+* Use the same cohort `sample_set_set` you created and used for `09-MergeBatchSites`.
+
+### Additional notes
+
+- The VCF produced by `15-CleanVcf` (and annotated by `17-AnnotateVcf`) prioritizes sensitivity, but additional downstream
+filtration is recommended to improve specificity.
+
diff --git a/website/docs/execution/overview.md b/website/docs/execution/overview.md
new file mode 100644
index 000000000..fecfd3825
--- /dev/null
+++ b/website/docs/execution/overview.md
@@ -0,0 +1,10 @@
+---
+title: Overview
+description: Overview
+sidebar_position: 1
+slug: overview
+---
+
+This section provides technical documentation for the running single-sample and joint calling modes in Terra.
+
+Please review the [Getting Started](/docs/gs/overview) material before proceeding with this section.
diff --git a/website/docs/execution/single.md b/website/docs/execution/single.md
new file mode 100644
index 000000000..1132a2ac3
--- /dev/null
+++ b/website/docs/execution/single.md
@@ -0,0 +1,121 @@
+---
+title: Single-sample
+description: Run the pipeline on a single sample
+sidebar_position: 3
+slug: single
+---
+
+## Introduction
+
+**Extending SV detection to small datasets**
+
+The Single Sample pipeline is designed to facilitate running the methods developed for the cohort-mode GATK-SV pipeline on small data sets or in
+clinical contexts where batching large numbers of samples is not an option. To do so, it uses precomputed data, SV calls,
+and model parameters computed by the cohort pipeline on a reference panel composed of similar samples. The pipeline integrates this
+precomputed information with signals extracted from the input CRAM file to produce a call set similar in quality to results
+computed by the cohort pipeline in a computationally tractable and reproducible manner.
+
+
+## Terra workspace
+Users should clone the [Terra single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+which is configured with a demo sample to run on.
+Refer to the following sections for instructions on how to run the pipeline on your data using this workspace.
+
+## Data
+
+### Case sample
+
+The default workspace includes a NA12878 input WGS CRAM file configured in the workspace samples data table. This file is part of the
+high coverage (30X) [WGS data](https://www.internationalgenome.org/data-portal/data-collection/30x-grch38) for the 1000
+Genomes Project samples generated by the New York Genome Center and hosted in
+[AnVIL](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019).
+
+### Reference panel
+
+The reference panel configured in the workspace consists of data and calls computed from 156 publicly available samples
+chosen from the NYGC/AnVIL 1000 Genomes high coverage data linked above.
+
+Inputs to the pipeline for the reference panel include:
+- A precomputed SV callset VCF, and joint-called depth-based CNV call files
+- Raw calls for the reference panel samples from Manta and WHAM
+- Trained models for calling copy number variation in GATK gCNV case mode
+- Parameters learned by the cohort mode pipeline in training machine learning models on the reference panel samples.
+
+These resources are primarily configured in the "Workspace Data" for this workspace. However, several of the resources need
+to be passed to the workflow as large lists of files or strings. Due to Terra limitations on uploading data containing lists to the
+workspace data table, these resources are specified directly in the workflow configuration.
+
+### Reference resources
+
+The pipeline uses a number of resource and data files computed for the hg38 reference:
+- Reference sequences and indices
+- Genome annotation tracks such as segmental duplication and RepeatMasker tracks
+- Data used for annotation of called variants, including GenCode gene annotations and gnomAD site allele frequencies.
+
+## Single sample workflow
+
+### What does it do?
+
+The workflow `gatk-sv-single-sample` calls structural variations on a single input CRAM by running the GATK-SV Single Sample Pipeline end-to-end.
+
+#### What does it require as input?
+
+The workflow accepts a single CRAM or BAM file as input, configured in the following parameters:
+
+|Input Type|Input Name|Description|
+|---------|--------|--------------|
+|`String`|`sample_id`|Case sample identifier|
+|`File`|`bam_or_cram_file`|Path to the GCS location of the input CRAM or BAM file|
+|`String`|`batch`|Arbitrary name to be assigned to the run|
+
+#### Additional workspace-level inputs
+
+- Reference resources for hg38
+- Input data for the reference panel
+- The set of docker images used in the pipeline.
+
+Please contact GATK-SV developers if you are interested in customizing these
+inputs beyond their defaults.
+
+#### What does it return as output?
+
+|Output Type|Output Name|Description|
+|---------|--------|--------------|
+|`File`|`final_vcf`|SV VCF output for the pipeline. Includes all sites genotyped as variant in the case sample and genotypes for the reference panel. Sites are annotated with overlap of functional genome elements and allele frequencies of matching variants in gnomAD|
+|`File`|`final_vcf_idx`|Index file for `final_vcf`|
+|`File`|`final_bed`|Final output in BED format. Filter status, list of variant samples, and all VCF INFO fields are reported as additional columns.|
+|`File`|`metrics_file`|Metrics computed from the input data and intermediate and final VCFs. Includes metrics on the SV evidence, and on the number of variants called, broken down by type and size range.|
+|`File`|`qc_file`|Quality-control check file. This extracts several key metrics from the `metrics_file` and compares them to pre-specified threshold values. If any QC checks evaluate to FAIL, further diagnostics may be required.|
+|`File`|`ploidy_matrix`|Matrix of contig ploidy estimates computed by GATK gCNV.|
+|`File`|`ploidy_plots`|Plots of contig ploidy generated from `ploidy_matrix`|
+|`File`|`non_genotyped_unique_depth_calls`|This VCF file contains any depth based calls made in the case sample that did not pass genotyping checks and do not match a depth-based call from the reference panel. If very high sensitivity is desired, examine this file for additional large CNV calls.|
+|`File`|`non_genotyped_unique_depth_calls_idx`|Index file for `non_genotyped_unique_depth_calls`|
+|`File`|`pre_cleanup_vcf`|VCF output in a representation used internally in the pipeline. This file is less compliant with the VCF spec and is intended for debugging purposes.|
+|`File`|`pre_cleanup_vcf_idx`|Index file for `pre_cleanup_vcf`|
+
+#### Example time and cost run on sample data
+
+|Sample Name|Sample Size|Time|Cost $|
+|-----------|-----------|----|------|
+|NA12878|18.17 GiB|23hrs|~$7.34|
+
+#### To use this workflow on your own data
+
+If you would like to run this workflow on your own samples (which must be medium-to-high coverage WGS data):
+
+- Clone the [workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample) into a Terra project you have access to
+- In the cloned workspace, upload rows to the Sample and (optionally) the Participant Data Table that describe your samples.
+ Ensure that the rows you add to the Sample table contain the columns `sample_id` and `bam_or_cram_file` are populated appropriately.
+- There is no need to modify values in the workspace data or method configuration. If you are interested in modifying the reference
+ genome resources or reference panel, please contact the GATK team for support as listed below.
+- Launch the workflow from the "Workflows" tab, selecting your samples as the inputs.
+
+#### Quality control
+
+Please check the `qc_file` output to screen for data quality issues. This file provides a table of quality metrics and
+suggested acceptable ranges.
+
+:::warning
+If a metric exceeds the recommended range, all variants will be automatically flagged with
+a non-passing `FILTER` status in the output VCF.
+:::
\ No newline at end of file
diff --git a/website/docs/gs/_category_.json b/website/docs/gs/_category_.json
index 6b9e1272a..fe28ad97e 100644
--- a/website/docs/gs/_category_.json
+++ b/website/docs/gs/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Getting Started",
- "position": 2,
+ "position": 3,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/gs/calling_modes.md b/website/docs/gs/calling_modes.md
new file mode 100644
index 000000000..dcafa5634
--- /dev/null
+++ b/website/docs/gs/calling_modes.md
@@ -0,0 +1,41 @@
+---
+title: Calling modes
+description: Description of single-sample and joint calling
+sidebar_position: 1
+---
+
+# Calling modes
+
+GATK-SV offers two different modes for SV calling. Users should carefully review the following sections to determine
+which mode is appropriate for their use case.
+
+## Single-sample mode
+
+GATK-SV can perform SV calling on individual samples. In this mode, a sample is jointly called against a fixed reference
+panel of [156 high-quality samples from the 1000 Genomes Project](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019). Single-sample mode is a good option for the following
+use cases:
+
+- Studies involving fewer 100 samples
+- Studies with rolling data delivery, i.e. in small batches over time
+
+Users should also consider that the single-sample mode is provided as a single workflow and is therefore considerably
+simpler to run than joint calling. However, it also has higher compute costs on a per-sample basis and will not be as sensitive
+as joint calling with larger cohorts.
+
+## Joint calling mode
+
+GATK-SV can also perform joint calling on a set of samples. Users may opt for this mode in the following use cases:
+
+- Studies involving at least 100 samples
+- When maximum sensitivity is desired
+- Data sets that are technically heterogeneous, i.e. with strong batch effects, or are very different from the single-sample mode reference panel
+
+Joint calling has the advantage of increasing SV discovery sensitivity and providing allele frequency estimates, and there are
+some features, such as genotype recalibration and filtering and in-depth QC plotting, that are only available in joint calling mode.
+However, this pipeline is considerably more complex to execute than the single-sample mode, requiring sample batching and the execution of
+several individual modules.
+
+## Related content
+
+More information on single-sample and joint calling can be found in the [Execution](/docs/execution/overview) section.
+
diff --git a/website/docs/gs/dockers.md b/website/docs/gs/dockers.md
new file mode 100644
index 000000000..7a4e0f658
--- /dev/null
+++ b/website/docs/gs/dockers.md
@@ -0,0 +1,73 @@
+---
+title: Docker images
+description: Docker images
+sidebar_position: 4
+---
+
+GATK-SV utilizes a set of [Docker](https://www.docker.com/) images for execution within containerized environments.
+
+### Publishing and availability
+
+Dockers are automatically built and pushed to the `us.gcr.io/broad-dsde-methods/gatk-sv` repository under two different conditions:
+1. **Release**: upon releasing a new version of GATK-SV. These Dockers are made permanently available.
+2. **Commit**: upon merging a new commit to the development branch. These Dockers are ephemeral and may be periodically
+deleted. Any users needing to preserve access to these Docker images should copy them to their own repository. Also
+note that these images are built on an "as-needed" basis, meaning an image is only updated if it has been changed
+in any way by the commit.
+
+The full set of current images are automatically published and pushed to a public
+[Google Artifact Registry](https://cloud.google.com/artifact-registry/docs) and listed in the
+[dockers.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers.json)
+file.
+
+:::info
+Microsoft Azure mirrors of all images can be found in
+[dockers_azure.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers_azure.json),
+but these are not currently available for public use.
+:::
+
+### Regions (IMPORTANT)
+
+Users using Google Compute VMs outside of the `us-central1` region, i.e. in other NA regions or continents, must copy all
+docker images to a repository hosted in their region. If using a Terra workspace, the region is listed under `Cloud
+Information`:`Location` in the workspace dashboard. Please see
+[this article](https://support.terra.bio/hc/en-us/articles/4408985788187-How-to-configure-Google-Artifact-Registry-to-prevent-data-transfer-egress-charges)
+for more information.
+
+:::warning
+Failure to localize Docker images to your region will incur significant egress costs.
+:::
+
+### Versioning
+
+All Docker images are tagged with a date and version number that must be run with the corresponding version of the
+WDLs. The Docker images built with a particular version can be determined from the `dockers.json` file by checking out
+the commit or release of interest and examining `dockers.json`, e.g.
+[v0.29-beta](https://github.com/broadinstitute/gatk-sv/blob/v0.29-beta/inputs/values/dockers.json).
+
+Note that a given commit may contain a mixture of Docker image version tags if only a subset of images has actually
+been updated, reflecting the "as-needed" rebuild policy described above.
+
+All Terra workspace releases have the WDLs and Docker images synchronized.
+
+:::info
+Cloning a Terra workspace copies a snapshot of the current workspace. Any future updates to the original workspace
+will not be propagated to the clone.
+:::
+
+:::warning
+We strongly recommend using a single version of GATK-SV for all projects. Switching versions may lead to batch effects
+and/or compromise call set fidelity.
+:::
+
+
+### Workflow inputs
+
+All GATK-SV WDLs utilize a subset of the Docker images, which are specified as inputs. For example,
+the `sv_pipeline` docker is supplied to the input `sv_pipeline_docker` in many WDLs.
+
+
+### Builds
+
+An in-depth guide to the GATK-SV Docker system and building Docker images is available in the
+[Advanced section](/docs/category/docker-builds).
diff --git a/website/docs/gs/input_files.md b/website/docs/gs/input_files.md
index 38e143869..aafd031f1 100644
--- a/website/docs/gs/input_files.md
+++ b/website/docs/gs/input_files.md
@@ -1,62 +1,72 @@
---
-title: Input Data
+title: Input data
description: Supported input and reference data.
-sidebar_position: 5
+sidebar_position: 2
slug: ./inputs
---
GATK-SV requires the following input data:
-- Illumina short-read whole-genome CRAMs or BAMs, aligned to hg38 with [bwa-mem](https://github.com/lh3/bwa).
- BAMs must also be indexed.
+1. Sequencing alignments in BAM or CRAM format that are:
+ - Short-read, paired-end Illumina (e.g. Novaseq)
+ - Deep whole-genome coverage (~30x); RNA-seq and targeted (exome) libraries are not supported
+ - Indexed (have a companion `.bai` or `.crai` file)
+ - Aligned to
+ [hg38](https://gatk.broadinstitute.org/hc/en-us/articles/360035890951-Human-genome-reference-builds-GRCh38-or-hg38-b37-hg19)
+ with either [GATK Best Practices](https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows)
+ and [bwa-mem](https://github.com/lh3/bwa),
+ or [Illumina DRAGEN](https://www.illumina.com/products/by-type/informatics-products/dragen-secondary-analysis.html) v3.4.12 or v3.7.8
+2. (Joint calling mode only) Family structure definitions file in [PED format](/docs/gs/inputs#ped-format). This file is required even if your dataset does not contain related individuals.
-- Family structure definitions file in
- [PED format](/docs/gs/inputs#ped-format).
-
-### PED file format {#ped-format}
-The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
-* The file must be tab-delimited.
-* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
-* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
-* Missing parental IDs should be entered as 0.
-* Header lines are allowed if they begin with a # character.
-To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
+Note that the supported alignment pipeline versions have been extensively tested for robustness and accuracy. While other
+versions of DRAGEN may work as well, they have not been validated with GATK-SV. We do not recommend mixing aligners within call sets.
### Sample Exclusion
We recommend filtering out samples with a high percentage
-of improperly paired reads (>10% or an outlier for your data)
+of improperly paired or chimeric reads
as technical outliers prior to running [GatherSampleEvidence](/docs/modules/gse).
-A high percentage of improperly paired reads may indicate issues
-with library prep, degradation, or contamination. Artifactual
-improperly paired reads could cause incorrect SV calls, and
-these samples have been observed to have longer runtimes and
-higher compute costs for [GatherSampleEvidence](/docs/modules/gse).
+Samples with high rates of anomalous reads may indicate issues
+with library preparation, degradation, or contamination and can lead to poor variant set quality.
+Samples failing these criteria often require longer run times and higher compute costs.
-### Sample ID requirements {#sampleids}
-#### Sample IDs must
+### Sample IDs {#sampleids}
+GATK-SV imposes certain restrictions on sample names (IDs) in order to avoid certain parsing errors (e.g. with the
+use of the `grep` command). While future releases will obviate some of these restrictions, users must modify
+their sample IDs according to the following requirements.
+
+#### Sample IDs must:
- Be unique within the cohort
- Contain only alphanumeric characters and underscores (no dashes, whitespace, or special characters)
-#### Sample IDs should not
-
-- Contain only numeric characters
+#### Sample IDs should not:
+- Contain only numeric characters, e.g. `10004928`
- Be a substring of another sample ID in the same cohort
- Contain any of the following substrings: `chr`, `name`, `DEL`, `DUP`, `CPX`, `CHROM`
-The same requirements apply to family IDs in the PED file,
-as well as batch IDs and the cohort ID provided as workflow inputs.
+The same requirements apply to family IDs in the PED file, as well as batch IDs and the cohort ID provided as workflow inputs.
-Sample IDs are provided to GatherSampleEvidence directly and
-need not match sample names from the BAM/CRAM headers.
-`GetSampleID.wdl` can be used to fetch BAM sample IDs and
-also generates a set of alternate IDs that are considered
-safe for this pipeline; alternatively, [this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
+Users should set sample IDs in [GatherSampleEvidence](/docs/modules/gse) with the `sample_id` input, which needs not match
+the sample name defined in the BAM/CRAM header. `GetSampleID.wdl` can be used to fetch BAM sample IDs and also generates a set
+of alternate IDs that are considered safe for this pipeline. Alternatively,
+[this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
transforms a list of sample IDs to fit these requirements.
-Currently, sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe).
-The following inputs will need to be updated with the transformed sample IDs:
+Sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe). To do so, set the parameter
+`rename_samples = True` and provide updated sample IDs via the `samples` parameter.
+
+Note that following inputs will need to be updated with the transformed sample IDs:
- Sample ID list for [GatherSampleEvidence](/docs/modules/gse) or [GatherBatchEvidence](/docs/modules/gbe)
- PED file
+
+
+### PED file format {#ped-format}
+The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
+* The file must be tab-delimited.
+* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
+* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
+* Missing parental IDs should be entered as 0.
+* Header lines are allowed if they begin with a # character.
+* To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
diff --git a/website/docs/gs/overview.md b/website/docs/gs/overview.md
index 5b147c973..f4493759c 100644
--- a/website/docs/gs/overview.md
+++ b/website/docs/gs/overview.md
@@ -6,9 +6,11 @@ sidebar_position: 0
# Overview
-GATK-SV is a highly-scalable cloud-native pipeline for structural variation discovery
-on Illumina short-read whole-genome sequencing (WGS) data.
-The pipeline genotypes structural variations using Docker-based tools, modularized in
-different components, and orchestrated using Cromwell.
+This section provides guidance on how GATK-SV can be used, restrictions on input data, and where to get started
+with generating call sets.
-Please refer to the documentation on [this page](/docs/run/overview) for running the pipeline.
+We highly recommend that users new to SV calling also review the [Best Practices Guide](/docs/best_practices).
+
+Most users should use the Terra platform for running GATK-SV and, after reading this section,
+should refer to the documentation in the [Execution section](/docs/execution/overview) for instructions on
+how to run their own data.
diff --git a/website/docs/gs/runtime_env.md b/website/docs/gs/runtime_env.md
index 65aee6032..cdc31f4ea 100644
--- a/website/docs/gs/runtime_env.md
+++ b/website/docs/gs/runtime_env.md
@@ -1,24 +1,30 @@
---
-title: Runtime Environments
+title: Runtime environments
description: Describes the supported runtime environments.
-sidebar_position: 7
+sidebar_position: 5
slug: ./runtime-env
---
-The GATK-SV pipeline consists of _workflows_ and _reference data_ that
-orchestrates the analysis flow of input data. Hence, a successful
-execution requires running the _workflows_ on _reference_ and input data.
+The GATK-SV pipeline consists of workflows implemented in the Workflow Description Language
+([WDL](https://openwdl.org/)) and is built for use on the Google Cloud Platform ([GCP](https://cloud.google.com/)).
-:::info Currently supported backends: GCP
-GATK-SV has been tested only on the Google Cloud Platform (GCP);
-therefore, we are unable to provide specific guidance or support
-for other execution platforms including HPC clusters and AWS.
-:::
+### Terra (recommended)
+To facilitate easy-of-use, security, and collaboration, GATK-SV is available on the [Terra](https://app.terra.bio/)
+platform. Users should clone pre-configured Terra workspaces as a starting point for running GATK-SV:
-## Alternative backends
+- [Single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+- Joint calling workspace (TODO)
-Contributions from the community to improve portability between backends
-will be considered on a case-by-case-basis. We ask contributors to
+These workspaces are actively maintained by the development team and will be updated with critical fixes and major releases.
+
+### Cromwell (advanced)
+Advanced users and developers who wish to run GATK-SV on a dedicated Cromwell server using GCP as a backend should refer
+to the [Advanced Guides](/docs/category/advanced-guides) section.
+
+### Alternative backends (not supported)
+
+Use of other backends (e.g. AWS or on-prem HPC clusters) is not currently supported. However, contributions from the
+community to improve portability between backends will be considered on a case-by-case-basis. We ask contributors to
please adhere to the following guidelines when submitting issues and pull requests:
1. Code changes must be functionally equivalent on GCP backends, i.e. not result in changed output
diff --git a/website/docs/gs/sv_callers.md b/website/docs/gs/sv_callers.md
new file mode 100644
index 000000000..165bf87b3
--- /dev/null
+++ b/website/docs/gs/sv_callers.md
@@ -0,0 +1,28 @@
+---
+title: SV/CNV callers
+description: Summary of SV discovery tools
+sidebar_position: 3
+---
+
+GATK-SV uses an ensemble of SV discovery tools to produce a raw call set and then integrates, filters, refines,
+and annotates the calls from these tools to produce a final call set.
+
+The SV calling tools, sometimes referred to as "PE/SR" tools, include:
+- [Manta](https://github.com/Illumina/manta)
+- [Wham](https://github.com/zeeev/wham)
+- [Scramble](https://github.com/GeneDx/scramble)
+
+Depth-based calling of copy number variants (CNVs) is performed by two tools:
+- [GATK-gCNV](https://github.com/broadinstitute/gatk)
+- [cn.MOPS](https://bioconductor.org/packages/release/bioc/html/cn.mops.html)
+
+While it is possible to omit individual tools from the pipeline, it is strongly recommended to use the default
+configuration that runs all of them.
+
+:::note
+As of 2024, most published joint call sets generated with GATK-SV used the tool MELT, a state-of-the-art mobile element
+insertion (MEI) detector, instead of Scramble. Due to licensing restrictions, we cannot provide a public docker image
+or reference panel VCFs for this algorithm. The version of the pipeline configured in the Terra workspaces does not run
+MELT or include MELT calls for the reference panel. However, the Scramble tool has replaced MELT as an MEI caller and
+should provide similar performance.
+:::
diff --git a/website/docs/intro.md b/website/docs/intro.md
index 46d0da8a7..413f28245 100644
--- a/website/docs/intro.md
+++ b/website/docs/intro.md
@@ -4,29 +4,29 @@ description: GATK-SV
sidebar_position: 1
---
-GATK-SV is a comprehensive, cloud-based ensemble pipeline to capture and annotate all
-classes of structural variants (SV) from whole genome sequencing (WGS). It can detect
+GATK-SV is a comprehensive, cloud-based ensemble pipeline for discovering and annotating all
+classes of structural variants (SV) from whole genome sequencing (WGS) data. It can detect
deletions, duplications, multi-allelic copy number variants, balanced inversions,
insertions, translocations, and a diverse spectrum of complex SV. Briefly, GATK-SV
-maximizes the sensitivity of SV discovery by harmonizing output from five tools
-(Manta, Wham, cnMOPS, GATK-gCNV, MELT). In order to reduce false positives, raw SV
+maximizes the sensitivity of SV discovery by harmonizing output from five tools:
+Manta, Wham, Scramble, cnMOPS, and GATK-gCNV. To minimize false positives, raw SVs
are adjudicated and re-genotyped from read evidence considering all potential
-sequencing evidence including anomalous paired-end (PE) reads, split reads (SR) crossing
-a breakpoint, normalized read-depth (RD), and B-allele frequencies (BAF). It also
-fully resolves 11 classes of complex SV based on their breakpoint signature. GATK-SV
-has been designed to be deployed in the Google Cloud Platform via the cromwell
-execution engine, which allows massively parallel scaling. Further details about
-GATK--SV can be found in [Collins et al. 2020, Nature](https://www.nature.com/articles/s41586-020-2287-8).
+sequencing evidence including anomalous paired-end (PE) reads, split reads (SR),
+read-depth (RD), and B-allele frequencies (BAF). It also fully resolves 16 classes of complex
+SVs composed of multiple breakpoints. GATK-SV is intended for use on the [Terra](https://app.terra.bio/)
+ platform.
+### Methods
-A high-level description of GATK-SV is available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022487952155-Structural-variant-SV-discovery).
+Further details about GATK-SV methods can be found in [Collins et al. 2020](https://www.nature.com/articles/s41586-020-2287-8).
-### Citation
+### GATK Best Practices
-Please cite the following publication:
+Additional guidance on running GATK-SV is also available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022653744283-GATK-Best-Practices-for-Structural-Variation-Discovery-on-Single-Samples).
-- [Collins, Brand, et al. 2020. "A structural variation reference for medical and population genetics." Nature 581, 444-451.](https://doi.org/10.1038/s41586-020-2287-8)
+### Where to go from here
-Additional references:
+This documentation includes instructions for running the pipeline, technical implementation details, troubleshooting
+information, and guides for advanced users who wish to work with the source code or rebuild the project.
-- [Werling et al. 2018. "An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder." Nature genetics 50.5, 727-736.](https://doi.org/10.1038/s41588-018-0107-y)
+We recommend new users continue to the [Getting Started overview](/docs/gs/overview.md).
diff --git a/website/docs/modules/_category_.json b/website/docs/modules/_category_.json
index bb1a47f82..8f985b25a 100644
--- a/website/docs/modules/_category_.json
+++ b/website/docs/modules/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Modules",
- "position": 6,
+ "position": 5,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/modules/annotate_vcf.md b/website/docs/modules/annotate_vcf.md
index f4d965b22..1ec30cc47 100644
--- a/website/docs/modules/annotate_vcf.md
+++ b/website/docs/modules/annotate_vcf.md
@@ -1,23 +1,101 @@
---
-title: AnnotateVCF
-description: Annotate VCF (work in progress)
-sidebar_position: 13
+title: AnnotateVcf
+description: Annotate VCF
+sidebar_position: 20
slug: av
---
-Add annotations, such as the inferred function and
-allele frequencies of variants, to final VCF.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+Adds annotations, such as the inferred function and allele frequencies of variants, to a VCF.
Annotations methods include:
+* Functional annotation - The GATK tool [SVAnnotate](https://gatk.broadinstitute.org/hc/en-us/articles/13832752531355-SVAnnotate)
+ is used to annotate SVs with inferred functional consequence on protein-coding regions, regulatory regions such as
+ UTR and promoters, and other non-coding elements.
+* Allele Frequency (`AF`) annotation - annotate SVs with their allele frequencies across all samples, and samples of
+ specific sex, as well as specific subpopulations.
+* Allele Frequency annotation with external callset - annotate SVs with the allele frequencies of their overlapping SVs
+ in another callset, eg. the gnomAD-SV reference callset.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ fg: FilterGenotypes
+ avcf: AnnotateVcf
+ fg --> avcf
+
+ class avcf thisModule
+ class fg inModules
+```
+
+### Inputs
+
+#### `vcf`
+Any SV VCF. Running on the [genotype filtered VCF](./fg#filtered_vcf) is recommended.
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `protein_coding_gtf`
+Coding transcript definitions, see [here](/docs/resources#protein_coding_gtf).
+
+####
+
+### 10-GenotypeBatch
+
+Read the full GenotypeBatch documentation [here](/docs/modules/gb).
+* Use the same `sample_set` definitions you used for `03-TrainGCNV` through `08-FilterBatchSamples`.
+
+### Steps 11-17
+
+Read the full documentation for [RegenotypeCNVs](/docs/modules/rgcnvs), [MakeCohortVcf](/docs/modules/cvcf) (which
+includes `CombineBatches`, `ResolveComplexVariants`, `GenotypeComplexVariants`, `CleanVcf`, `MainVcfQc`), and
+[AnnotateVcf](/docs/modules/av).
+* Use the same cohort `sample_set_set` you created and used for `09-MergeBatchSites`.
+
+### Additional notes
+
+- The VCF produced by `15-CleanVcf` (and annotated by `17-AnnotateVcf`) prioritizes sensitivity, but additional downstream
+filtration is recommended to improve specificity.
+
diff --git a/website/docs/execution/overview.md b/website/docs/execution/overview.md
new file mode 100644
index 000000000..fecfd3825
--- /dev/null
+++ b/website/docs/execution/overview.md
@@ -0,0 +1,10 @@
+---
+title: Overview
+description: Overview
+sidebar_position: 1
+slug: overview
+---
+
+This section provides technical documentation for the running single-sample and joint calling modes in Terra.
+
+Please review the [Getting Started](/docs/gs/overview) material before proceeding with this section.
diff --git a/website/docs/execution/single.md b/website/docs/execution/single.md
new file mode 100644
index 000000000..1132a2ac3
--- /dev/null
+++ b/website/docs/execution/single.md
@@ -0,0 +1,121 @@
+---
+title: Single-sample
+description: Run the pipeline on a single sample
+sidebar_position: 3
+slug: single
+---
+
+## Introduction
+
+**Extending SV detection to small datasets**
+
+The Single Sample pipeline is designed to facilitate running the methods developed for the cohort-mode GATK-SV pipeline on small data sets or in
+clinical contexts where batching large numbers of samples is not an option. To do so, it uses precomputed data, SV calls,
+and model parameters computed by the cohort pipeline on a reference panel composed of similar samples. The pipeline integrates this
+precomputed information with signals extracted from the input CRAM file to produce a call set similar in quality to results
+computed by the cohort pipeline in a computationally tractable and reproducible manner.
+
+
+## Terra workspace
+Users should clone the [Terra single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+which is configured with a demo sample to run on.
+Refer to the following sections for instructions on how to run the pipeline on your data using this workspace.
+
+## Data
+
+### Case sample
+
+The default workspace includes a NA12878 input WGS CRAM file configured in the workspace samples data table. This file is part of the
+high coverage (30X) [WGS data](https://www.internationalgenome.org/data-portal/data-collection/30x-grch38) for the 1000
+Genomes Project samples generated by the New York Genome Center and hosted in
+[AnVIL](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019).
+
+### Reference panel
+
+The reference panel configured in the workspace consists of data and calls computed from 156 publicly available samples
+chosen from the NYGC/AnVIL 1000 Genomes high coverage data linked above.
+
+Inputs to the pipeline for the reference panel include:
+- A precomputed SV callset VCF, and joint-called depth-based CNV call files
+- Raw calls for the reference panel samples from Manta and WHAM
+- Trained models for calling copy number variation in GATK gCNV case mode
+- Parameters learned by the cohort mode pipeline in training machine learning models on the reference panel samples.
+
+These resources are primarily configured in the "Workspace Data" for this workspace. However, several of the resources need
+to be passed to the workflow as large lists of files or strings. Due to Terra limitations on uploading data containing lists to the
+workspace data table, these resources are specified directly in the workflow configuration.
+
+### Reference resources
+
+The pipeline uses a number of resource and data files computed for the hg38 reference:
+- Reference sequences and indices
+- Genome annotation tracks such as segmental duplication and RepeatMasker tracks
+- Data used for annotation of called variants, including GenCode gene annotations and gnomAD site allele frequencies.
+
+## Single sample workflow
+
+### What does it do?
+
+The workflow `gatk-sv-single-sample` calls structural variations on a single input CRAM by running the GATK-SV Single Sample Pipeline end-to-end.
+
+#### What does it require as input?
+
+The workflow accepts a single CRAM or BAM file as input, configured in the following parameters:
+
+|Input Type|Input Name|Description|
+|---------|--------|--------------|
+|`String`|`sample_id`|Case sample identifier|
+|`File`|`bam_or_cram_file`|Path to the GCS location of the input CRAM or BAM file|
+|`String`|`batch`|Arbitrary name to be assigned to the run|
+
+#### Additional workspace-level inputs
+
+- Reference resources for hg38
+- Input data for the reference panel
+- The set of docker images used in the pipeline.
+
+Please contact GATK-SV developers if you are interested in customizing these
+inputs beyond their defaults.
+
+#### What does it return as output?
+
+|Output Type|Output Name|Description|
+|---------|--------|--------------|
+|`File`|`final_vcf`|SV VCF output for the pipeline. Includes all sites genotyped as variant in the case sample and genotypes for the reference panel. Sites are annotated with overlap of functional genome elements and allele frequencies of matching variants in gnomAD|
+|`File`|`final_vcf_idx`|Index file for `final_vcf`|
+|`File`|`final_bed`|Final output in BED format. Filter status, list of variant samples, and all VCF INFO fields are reported as additional columns.|
+|`File`|`metrics_file`|Metrics computed from the input data and intermediate and final VCFs. Includes metrics on the SV evidence, and on the number of variants called, broken down by type and size range.|
+|`File`|`qc_file`|Quality-control check file. This extracts several key metrics from the `metrics_file` and compares them to pre-specified threshold values. If any QC checks evaluate to FAIL, further diagnostics may be required.|
+|`File`|`ploidy_matrix`|Matrix of contig ploidy estimates computed by GATK gCNV.|
+|`File`|`ploidy_plots`|Plots of contig ploidy generated from `ploidy_matrix`|
+|`File`|`non_genotyped_unique_depth_calls`|This VCF file contains any depth based calls made in the case sample that did not pass genotyping checks and do not match a depth-based call from the reference panel. If very high sensitivity is desired, examine this file for additional large CNV calls.|
+|`File`|`non_genotyped_unique_depth_calls_idx`|Index file for `non_genotyped_unique_depth_calls`|
+|`File`|`pre_cleanup_vcf`|VCF output in a representation used internally in the pipeline. This file is less compliant with the VCF spec and is intended for debugging purposes.|
+|`File`|`pre_cleanup_vcf_idx`|Index file for `pre_cleanup_vcf`|
+
+#### Example time and cost run on sample data
+
+|Sample Name|Sample Size|Time|Cost $|
+|-----------|-----------|----|------|
+|NA12878|18.17 GiB|23hrs|~$7.34|
+
+#### To use this workflow on your own data
+
+If you would like to run this workflow on your own samples (which must be medium-to-high coverage WGS data):
+
+- Clone the [workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample) into a Terra project you have access to
+- In the cloned workspace, upload rows to the Sample and (optionally) the Participant Data Table that describe your samples.
+ Ensure that the rows you add to the Sample table contain the columns `sample_id` and `bam_or_cram_file` are populated appropriately.
+- There is no need to modify values in the workspace data or method configuration. If you are interested in modifying the reference
+ genome resources or reference panel, please contact the GATK team for support as listed below.
+- Launch the workflow from the "Workflows" tab, selecting your samples as the inputs.
+
+#### Quality control
+
+Please check the `qc_file` output to screen for data quality issues. This file provides a table of quality metrics and
+suggested acceptable ranges.
+
+:::warning
+If a metric exceeds the recommended range, all variants will be automatically flagged with
+a non-passing `FILTER` status in the output VCF.
+:::
\ No newline at end of file
diff --git a/website/docs/gs/_category_.json b/website/docs/gs/_category_.json
index 6b9e1272a..fe28ad97e 100644
--- a/website/docs/gs/_category_.json
+++ b/website/docs/gs/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Getting Started",
- "position": 2,
+ "position": 3,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/gs/calling_modes.md b/website/docs/gs/calling_modes.md
new file mode 100644
index 000000000..dcafa5634
--- /dev/null
+++ b/website/docs/gs/calling_modes.md
@@ -0,0 +1,41 @@
+---
+title: Calling modes
+description: Description of single-sample and joint calling
+sidebar_position: 1
+---
+
+# Calling modes
+
+GATK-SV offers two different modes for SV calling. Users should carefully review the following sections to determine
+which mode is appropriate for their use case.
+
+## Single-sample mode
+
+GATK-SV can perform SV calling on individual samples. In this mode, a sample is jointly called against a fixed reference
+panel of [156 high-quality samples from the 1000 Genomes Project](https://app.terra.bio/#workspaces/anvil-datastorage/1000G-high-coverage-2019). Single-sample mode is a good option for the following
+use cases:
+
+- Studies involving fewer 100 samples
+- Studies with rolling data delivery, i.e. in small batches over time
+
+Users should also consider that the single-sample mode is provided as a single workflow and is therefore considerably
+simpler to run than joint calling. However, it also has higher compute costs on a per-sample basis and will not be as sensitive
+as joint calling with larger cohorts.
+
+## Joint calling mode
+
+GATK-SV can also perform joint calling on a set of samples. Users may opt for this mode in the following use cases:
+
+- Studies involving at least 100 samples
+- When maximum sensitivity is desired
+- Data sets that are technically heterogeneous, i.e. with strong batch effects, or are very different from the single-sample mode reference panel
+
+Joint calling has the advantage of increasing SV discovery sensitivity and providing allele frequency estimates, and there are
+some features, such as genotype recalibration and filtering and in-depth QC plotting, that are only available in joint calling mode.
+However, this pipeline is considerably more complex to execute than the single-sample mode, requiring sample batching and the execution of
+several individual modules.
+
+## Related content
+
+More information on single-sample and joint calling can be found in the [Execution](/docs/execution/overview) section.
+
diff --git a/website/docs/gs/dockers.md b/website/docs/gs/dockers.md
new file mode 100644
index 000000000..7a4e0f658
--- /dev/null
+++ b/website/docs/gs/dockers.md
@@ -0,0 +1,73 @@
+---
+title: Docker images
+description: Docker images
+sidebar_position: 4
+---
+
+GATK-SV utilizes a set of [Docker](https://www.docker.com/) images for execution within containerized environments.
+
+### Publishing and availability
+
+Dockers are automatically built and pushed to the `us.gcr.io/broad-dsde-methods/gatk-sv` repository under two different conditions:
+1. **Release**: upon releasing a new version of GATK-SV. These Dockers are made permanently available.
+2. **Commit**: upon merging a new commit to the development branch. These Dockers are ephemeral and may be periodically
+deleted. Any users needing to preserve access to these Docker images should copy them to their own repository. Also
+note that these images are built on an "as-needed" basis, meaning an image is only updated if it has been changed
+in any way by the commit.
+
+The full set of current images are automatically published and pushed to a public
+[Google Artifact Registry](https://cloud.google.com/artifact-registry/docs) and listed in the
+[dockers.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers.json)
+file.
+
+:::info
+Microsoft Azure mirrors of all images can be found in
+[dockers_azure.json](https://github.com/broadinstitute/gatk-sv/blob/main/inputs/values/dockers_azure.json),
+but these are not currently available for public use.
+:::
+
+### Regions (IMPORTANT)
+
+Users using Google Compute VMs outside of the `us-central1` region, i.e. in other NA regions or continents, must copy all
+docker images to a repository hosted in their region. If using a Terra workspace, the region is listed under `Cloud
+Information`:`Location` in the workspace dashboard. Please see
+[this article](https://support.terra.bio/hc/en-us/articles/4408985788187-How-to-configure-Google-Artifact-Registry-to-prevent-data-transfer-egress-charges)
+for more information.
+
+:::warning
+Failure to localize Docker images to your region will incur significant egress costs.
+:::
+
+### Versioning
+
+All Docker images are tagged with a date and version number that must be run with the corresponding version of the
+WDLs. The Docker images built with a particular version can be determined from the `dockers.json` file by checking out
+the commit or release of interest and examining `dockers.json`, e.g.
+[v0.29-beta](https://github.com/broadinstitute/gatk-sv/blob/v0.29-beta/inputs/values/dockers.json).
+
+Note that a given commit may contain a mixture of Docker image version tags if only a subset of images has actually
+been updated, reflecting the "as-needed" rebuild policy described above.
+
+All Terra workspace releases have the WDLs and Docker images synchronized.
+
+:::info
+Cloning a Terra workspace copies a snapshot of the current workspace. Any future updates to the original workspace
+will not be propagated to the clone.
+:::
+
+:::warning
+We strongly recommend using a single version of GATK-SV for all projects. Switching versions may lead to batch effects
+and/or compromise call set fidelity.
+:::
+
+
+### Workflow inputs
+
+All GATK-SV WDLs utilize a subset of the Docker images, which are specified as inputs. For example,
+the `sv_pipeline` docker is supplied to the input `sv_pipeline_docker` in many WDLs.
+
+
+### Builds
+
+An in-depth guide to the GATK-SV Docker system and building Docker images is available in the
+[Advanced section](/docs/category/docker-builds).
diff --git a/website/docs/gs/input_files.md b/website/docs/gs/input_files.md
index 38e143869..aafd031f1 100644
--- a/website/docs/gs/input_files.md
+++ b/website/docs/gs/input_files.md
@@ -1,62 +1,72 @@
---
-title: Input Data
+title: Input data
description: Supported input and reference data.
-sidebar_position: 5
+sidebar_position: 2
slug: ./inputs
---
GATK-SV requires the following input data:
-- Illumina short-read whole-genome CRAMs or BAMs, aligned to hg38 with [bwa-mem](https://github.com/lh3/bwa).
- BAMs must also be indexed.
+1. Sequencing alignments in BAM or CRAM format that are:
+ - Short-read, paired-end Illumina (e.g. Novaseq)
+ - Deep whole-genome coverage (~30x); RNA-seq and targeted (exome) libraries are not supported
+ - Indexed (have a companion `.bai` or `.crai` file)
+ - Aligned to
+ [hg38](https://gatk.broadinstitute.org/hc/en-us/articles/360035890951-Human-genome-reference-builds-GRCh38-or-hg38-b37-hg19)
+ with either [GATK Best Practices](https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows)
+ and [bwa-mem](https://github.com/lh3/bwa),
+ or [Illumina DRAGEN](https://www.illumina.com/products/by-type/informatics-products/dragen-secondary-analysis.html) v3.4.12 or v3.7.8
+2. (Joint calling mode only) Family structure definitions file in [PED format](/docs/gs/inputs#ped-format). This file is required even if your dataset does not contain related individuals.
-- Family structure definitions file in
- [PED format](/docs/gs/inputs#ped-format).
-
-### PED file format {#ped-format}
-The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
-* The file must be tab-delimited.
-* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
-* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
-* Missing parental IDs should be entered as 0.
-* Header lines are allowed if they begin with a # character.
-To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
+Note that the supported alignment pipeline versions have been extensively tested for robustness and accuracy. While other
+versions of DRAGEN may work as well, they have not been validated with GATK-SV. We do not recommend mixing aligners within call sets.
### Sample Exclusion
We recommend filtering out samples with a high percentage
-of improperly paired reads (>10% or an outlier for your data)
+of improperly paired or chimeric reads
as technical outliers prior to running [GatherSampleEvidence](/docs/modules/gse).
-A high percentage of improperly paired reads may indicate issues
-with library prep, degradation, or contamination. Artifactual
-improperly paired reads could cause incorrect SV calls, and
-these samples have been observed to have longer runtimes and
-higher compute costs for [GatherSampleEvidence](/docs/modules/gse).
+Samples with high rates of anomalous reads may indicate issues
+with library preparation, degradation, or contamination and can lead to poor variant set quality.
+Samples failing these criteria often require longer run times and higher compute costs.
-### Sample ID requirements {#sampleids}
-#### Sample IDs must
+### Sample IDs {#sampleids}
+GATK-SV imposes certain restrictions on sample names (IDs) in order to avoid certain parsing errors (e.g. with the
+use of the `grep` command). While future releases will obviate some of these restrictions, users must modify
+their sample IDs according to the following requirements.
+
+#### Sample IDs must:
- Be unique within the cohort
- Contain only alphanumeric characters and underscores (no dashes, whitespace, or special characters)
-#### Sample IDs should not
-
-- Contain only numeric characters
+#### Sample IDs should not:
+- Contain only numeric characters, e.g. `10004928`
- Be a substring of another sample ID in the same cohort
- Contain any of the following substrings: `chr`, `name`, `DEL`, `DUP`, `CPX`, `CHROM`
-The same requirements apply to family IDs in the PED file,
-as well as batch IDs and the cohort ID provided as workflow inputs.
+The same requirements apply to family IDs in the PED file, as well as batch IDs and the cohort ID provided as workflow inputs.
-Sample IDs are provided to GatherSampleEvidence directly and
-need not match sample names from the BAM/CRAM headers.
-`GetSampleID.wdl` can be used to fetch BAM sample IDs and
-also generates a set of alternate IDs that are considered
-safe for this pipeline; alternatively, [this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
+Users should set sample IDs in [GatherSampleEvidence](/docs/modules/gse) with the `sample_id` input, which needs not match
+the sample name defined in the BAM/CRAM header. `GetSampleID.wdl` can be used to fetch BAM sample IDs and also generates a set
+of alternate IDs that are considered safe for this pipeline. Alternatively,
+[this script](https://github.com/talkowski-lab/gnomad_sv_v3/blob/master/sample_id/convert_sample_ids.py)
transforms a list of sample IDs to fit these requirements.
-Currently, sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe).
-The following inputs will need to be updated with the transformed sample IDs:
+Sample IDs can be replaced again in [GatherBatchEvidence](/docs/modules/gbe). To do so, set the parameter
+`rename_samples = True` and provide updated sample IDs via the `samples` parameter.
+
+Note that following inputs will need to be updated with the transformed sample IDs:
- Sample ID list for [GatherSampleEvidence](/docs/modules/gse) or [GatherBatchEvidence](/docs/modules/gbe)
- PED file
+
+
+### PED file format {#ped-format}
+The PED file format is described [here](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format). Note that GATK-SV imposes additional requirements:
+* The file must be tab-delimited.
+* The sex column must only contain 0, 1, or 2: 1=Male, 2=Female, 0=Other/Unknown. Sex chromosome aneuploidies (detected in [EvidenceQC](/docs/modules/eqc)) should be entered as sex = 0.
+* All family, individual, and parental IDs must conform to the [sample ID requirements](/docs/gs/inputs#sampleids).
+* Missing parental IDs should be entered as 0.
+* Header lines are allowed if they begin with a # character.
+* To validate the PED file, you may use `src/sv-pipeline/scripts/validate_ped.py -p pedigree.ped -s samples.list`.
diff --git a/website/docs/gs/overview.md b/website/docs/gs/overview.md
index 5b147c973..f4493759c 100644
--- a/website/docs/gs/overview.md
+++ b/website/docs/gs/overview.md
@@ -6,9 +6,11 @@ sidebar_position: 0
# Overview
-GATK-SV is a highly-scalable cloud-native pipeline for structural variation discovery
-on Illumina short-read whole-genome sequencing (WGS) data.
-The pipeline genotypes structural variations using Docker-based tools, modularized in
-different components, and orchestrated using Cromwell.
+This section provides guidance on how GATK-SV can be used, restrictions on input data, and where to get started
+with generating call sets.
-Please refer to the documentation on [this page](/docs/run/overview) for running the pipeline.
+We highly recommend that users new to SV calling also review the [Best Practices Guide](/docs/best_practices).
+
+Most users should use the Terra platform for running GATK-SV and, after reading this section,
+should refer to the documentation in the [Execution section](/docs/execution/overview) for instructions on
+how to run their own data.
diff --git a/website/docs/gs/runtime_env.md b/website/docs/gs/runtime_env.md
index 65aee6032..cdc31f4ea 100644
--- a/website/docs/gs/runtime_env.md
+++ b/website/docs/gs/runtime_env.md
@@ -1,24 +1,30 @@
---
-title: Runtime Environments
+title: Runtime environments
description: Describes the supported runtime environments.
-sidebar_position: 7
+sidebar_position: 5
slug: ./runtime-env
---
-The GATK-SV pipeline consists of _workflows_ and _reference data_ that
-orchestrates the analysis flow of input data. Hence, a successful
-execution requires running the _workflows_ on _reference_ and input data.
+The GATK-SV pipeline consists of workflows implemented in the Workflow Description Language
+([WDL](https://openwdl.org/)) and is built for use on the Google Cloud Platform ([GCP](https://cloud.google.com/)).
-:::info Currently supported backends: GCP
-GATK-SV has been tested only on the Google Cloud Platform (GCP);
-therefore, we are unable to provide specific guidance or support
-for other execution platforms including HPC clusters and AWS.
-:::
+### Terra (recommended)
+To facilitate easy-of-use, security, and collaboration, GATK-SV is available on the [Terra](https://app.terra.bio/)
+platform. Users should clone pre-configured Terra workspaces as a starting point for running GATK-SV:
-## Alternative backends
+- [Single-sample workspace](https://app.terra.bio/#workspaces/help-gatk/GATK-Structural-Variants-Single-Sample)
+- Joint calling workspace (TODO)
-Contributions from the community to improve portability between backends
-will be considered on a case-by-case-basis. We ask contributors to
+These workspaces are actively maintained by the development team and will be updated with critical fixes and major releases.
+
+### Cromwell (advanced)
+Advanced users and developers who wish to run GATK-SV on a dedicated Cromwell server using GCP as a backend should refer
+to the [Advanced Guides](/docs/category/advanced-guides) section.
+
+### Alternative backends (not supported)
+
+Use of other backends (e.g. AWS or on-prem HPC clusters) is not currently supported. However, contributions from the
+community to improve portability between backends will be considered on a case-by-case-basis. We ask contributors to
please adhere to the following guidelines when submitting issues and pull requests:
1. Code changes must be functionally equivalent on GCP backends, i.e. not result in changed output
diff --git a/website/docs/gs/sv_callers.md b/website/docs/gs/sv_callers.md
new file mode 100644
index 000000000..165bf87b3
--- /dev/null
+++ b/website/docs/gs/sv_callers.md
@@ -0,0 +1,28 @@
+---
+title: SV/CNV callers
+description: Summary of SV discovery tools
+sidebar_position: 3
+---
+
+GATK-SV uses an ensemble of SV discovery tools to produce a raw call set and then integrates, filters, refines,
+and annotates the calls from these tools to produce a final call set.
+
+The SV calling tools, sometimes referred to as "PE/SR" tools, include:
+- [Manta](https://github.com/Illumina/manta)
+- [Wham](https://github.com/zeeev/wham)
+- [Scramble](https://github.com/GeneDx/scramble)
+
+Depth-based calling of copy number variants (CNVs) is performed by two tools:
+- [GATK-gCNV](https://github.com/broadinstitute/gatk)
+- [cn.MOPS](https://bioconductor.org/packages/release/bioc/html/cn.mops.html)
+
+While it is possible to omit individual tools from the pipeline, it is strongly recommended to use the default
+configuration that runs all of them.
+
+:::note
+As of 2024, most published joint call sets generated with GATK-SV used the tool MELT, a state-of-the-art mobile element
+insertion (MEI) detector, instead of Scramble. Due to licensing restrictions, we cannot provide a public docker image
+or reference panel VCFs for this algorithm. The version of the pipeline configured in the Terra workspaces does not run
+MELT or include MELT calls for the reference panel. However, the Scramble tool has replaced MELT as an MEI caller and
+should provide similar performance.
+:::
diff --git a/website/docs/intro.md b/website/docs/intro.md
index 46d0da8a7..413f28245 100644
--- a/website/docs/intro.md
+++ b/website/docs/intro.md
@@ -4,29 +4,29 @@ description: GATK-SV
sidebar_position: 1
---
-GATK-SV is a comprehensive, cloud-based ensemble pipeline to capture and annotate all
-classes of structural variants (SV) from whole genome sequencing (WGS). It can detect
+GATK-SV is a comprehensive, cloud-based ensemble pipeline for discovering and annotating all
+classes of structural variants (SV) from whole genome sequencing (WGS) data. It can detect
deletions, duplications, multi-allelic copy number variants, balanced inversions,
insertions, translocations, and a diverse spectrum of complex SV. Briefly, GATK-SV
-maximizes the sensitivity of SV discovery by harmonizing output from five tools
-(Manta, Wham, cnMOPS, GATK-gCNV, MELT). In order to reduce false positives, raw SV
+maximizes the sensitivity of SV discovery by harmonizing output from five tools:
+Manta, Wham, Scramble, cnMOPS, and GATK-gCNV. To minimize false positives, raw SVs
are adjudicated and re-genotyped from read evidence considering all potential
-sequencing evidence including anomalous paired-end (PE) reads, split reads (SR) crossing
-a breakpoint, normalized read-depth (RD), and B-allele frequencies (BAF). It also
-fully resolves 11 classes of complex SV based on their breakpoint signature. GATK-SV
-has been designed to be deployed in the Google Cloud Platform via the cromwell
-execution engine, which allows massively parallel scaling. Further details about
-GATK--SV can be found in [Collins et al. 2020, Nature](https://www.nature.com/articles/s41586-020-2287-8).
+sequencing evidence including anomalous paired-end (PE) reads, split reads (SR),
+read-depth (RD), and B-allele frequencies (BAF). It also fully resolves 16 classes of complex
+SVs composed of multiple breakpoints. GATK-SV is intended for use on the [Terra](https://app.terra.bio/)
+ platform.
+### Methods
-A high-level description of GATK-SV is available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022487952155-Structural-variant-SV-discovery).
+Further details about GATK-SV methods can be found in [Collins et al. 2020](https://www.nature.com/articles/s41586-020-2287-8).
-### Citation
+### GATK Best Practices
-Please cite the following publication:
+Additional guidance on running GATK-SV is also available [here](https://gatk.broadinstitute.org/hc/en-us/articles/9022653744283-GATK-Best-Practices-for-Structural-Variation-Discovery-on-Single-Samples).
-- [Collins, Brand, et al. 2020. "A structural variation reference for medical and population genetics." Nature 581, 444-451.](https://doi.org/10.1038/s41586-020-2287-8)
+### Where to go from here
-Additional references:
+This documentation includes instructions for running the pipeline, technical implementation details, troubleshooting
+information, and guides for advanced users who wish to work with the source code or rebuild the project.
-- [Werling et al. 2018. "An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder." Nature genetics 50.5, 727-736.](https://doi.org/10.1038/s41588-018-0107-y)
+We recommend new users continue to the [Getting Started overview](/docs/gs/overview.md).
diff --git a/website/docs/modules/_category_.json b/website/docs/modules/_category_.json
index bb1a47f82..8f985b25a 100644
--- a/website/docs/modules/_category_.json
+++ b/website/docs/modules/_category_.json
@@ -1,6 +1,6 @@
{
"label": "Modules",
- "position": 6,
+ "position": 5,
"link": {
"type": "generated-index"
}
diff --git a/website/docs/modules/annotate_vcf.md b/website/docs/modules/annotate_vcf.md
index f4d965b22..1ec30cc47 100644
--- a/website/docs/modules/annotate_vcf.md
+++ b/website/docs/modules/annotate_vcf.md
@@ -1,23 +1,101 @@
---
-title: AnnotateVCF
-description: Annotate VCF (work in progress)
-sidebar_position: 13
+title: AnnotateVcf
+description: Annotate VCF
+sidebar_position: 20
slug: av
---
-Add annotations, such as the inferred function and
-allele frequencies of variants, to final VCF.
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+Adds annotations, such as the inferred function and allele frequencies of variants, to a VCF.
Annotations methods include:
+* Functional annotation - The GATK tool [SVAnnotate](https://gatk.broadinstitute.org/hc/en-us/articles/13832752531355-SVAnnotate)
+ is used to annotate SVs with inferred functional consequence on protein-coding regions, regulatory regions such as
+ UTR and promoters, and other non-coding elements.
+* Allele Frequency (`AF`) annotation - annotate SVs with their allele frequencies across all samples, and samples of
+ specific sex, as well as specific subpopulations.
+* Allele Frequency annotation with external callset - annotate SVs with the allele frequencies of their overlapping SVs
+ in another callset, eg. the gnomAD-SV reference callset.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ fg: FilterGenotypes
+ avcf: AnnotateVcf
+ fg --> avcf
+
+ class avcf thisModule
+ class fg inModules
+```
+
+### Inputs
+
+#### `vcf`
+Any SV VCF. Running on the [genotype filtered VCF](./fg#filtered_vcf) is recommended.
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. May be alphanumeric with underscores.
+
+#### `protein_coding_gtf`
+Coding transcript definitions, see [here](/docs/resources#protein_coding_gtf).
+
+####  -- **GatherBatchEvidence** Copy number variant calling using
- `cn.MOPS` and `GATK gCNV`; B-allele frequency (BAF) generation;
- call and evidence aggregation.
+Each module is implemented in the [Workflow Description Language (WDL)](https://openwdl.org). The Terra workspaces come
+pre-configured with default values for all required parameters and set up to run the pipeline for most use cases.
-- **ClusterBatch** Variant clustering
+The following sections supplement the Terra workspaces with documentation for each WDL, including an overview of its
+function, key input parameters, and outputs. Not all parameters are documented here, as some WDLs contain dozens of
+inputs. Descriptions of some common inputs can be found in the [Resource files](/docs/resources) and
+[Runtime attributes](/docs/runtime_attr) sections. Users are encouraged to refer to the WDL source code for additional
+clarification.
-- **GenerateBatchMetrics** Variant filtering metric generation
-
-- **FilterBatch** Variant filtering; outlier exclusion
-
-- **GenotypeBatch** Genotyping
-
-- **MakeCohortVcf** Cross-batch integration; complex variant resolution and re-genotyping; vcf cleanup
-
-- **Module 07 (in development)** Downstream filtering, including minGQ, batch effect check,
- outlier samples removal and final recalibration;
-
-- **AnnotateVCF** Annotations, including functional annotation,
- allele frequency (AF) annotation and AF annotation with external population callsets;
-
-- **Module 09 (in development)** Visualization, including scripts that generates IGV screenshots and rd plots.
-
-- Additional modules to be added: de novo and mosaic scripts
-
-
-## Pipeline Parameters
-
-Several inputs are shared across different modules of the pipeline, which are explained in this section.
-
-#### `ped_file`
-
-A pedigree file describing the familial relationships between the samples in the cohort.
-The file needs to be in the
-[PED format](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format).
-Updated with [EvidenceQC](./eqc) sex assignments, including
-`sex = 0` for sex aneuploidies;
-genotypes on chrX and chrY for samples with `sex = 0` in the PED file will be set to
-`./.` and these samples will be excluded from sex-specific training steps.
+For details on running GATK-SV on Terra, refer to the [Execution](/docs/execution/joint#instructions) section.
diff --git a/website/docs/modules/join_raw_calls.md b/website/docs/modules/join_raw_calls.md
new file mode 100644
index 000000000..55b563a24
--- /dev/null
+++ b/website/docs/modules/join_raw_calls.md
@@ -0,0 +1,54 @@
+---
+title: JoinRawCalls
+description: Clusters unfiltered variants across batches
+sidebar_position: 17
+slug: jrc
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/JoinRawCalls.wdl)
+
+This module clusters raw unfiltered variants from [ClusterBatch](./cb) across all batches. Concordance between these
+genotypes and the joint call set usually can be indicative of variant quality and is used downstream for genotype
+filtering.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cb: ClusterBatch
+ jrc: JoinRawCalls
+ svc: SVConcordance
+ cb --> jrc
+ jrc --> svc
+
+ class jrc thisModule
+ class cb inModules
+ class svc outModules
+```
+
+### Inputs
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. The guidelines outlined in the
+[sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `clustered_*_vcfs`
+Array of clustered VCFs for all batches, generated in [ClusterBatch](./cb#clustered__vcf).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+### Outputs
+
+#### `joined_raw_calls_vcf`
+VCF containing all raw calls in the cohort.
+
+#### `ploidy_table`
+TSV of contig ploidies for all samples, assuming diploid autosome and sex assignments from the [ped_file](#ped_file).
diff --git a/website/docs/modules/main_vcf_qc.md b/website/docs/modules/main_vcf_qc.md
new file mode 100644
index 000000000..d362765b4
--- /dev/null
+++ b/website/docs/modules/main_vcf_qc.md
@@ -0,0 +1,228 @@
+---
+title: MainVcfQC
+description: VCF QC plotting
+sidebar_position: 21
+slug: mvqc
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MainVcfQC.wdl)
+
+Creates plots for call set analysis and quality control (QC). This module is run by default with the
+[FilterGenotypes](./fg) module. Note, however, that the module is a stand-alone workflow and can be used with any batch-
+or cohort-level (i.e. multi-sample) SV VCF, including outputs from [ClusterBatch](./cb) and onward.
+
+Users can optionally provide external benchmarking datasets with the
+[site_level_comparison_datasets](#optional-site_level_comparison_datasets) and
+[sample_level_comparison_datasets](#optional-sample_level_comparison_datasets) parameters.
+
+The following sections provide guidance on interpreting these plots for VCFs that have been run with the recommended
+pipeline through [FilterGenotypes](./fg).
+
+### Output structure
+
+The tarball output has the following directory structure:
+
+- `/plots/main_plots` : main QC plots, described in detail [below](#plots-description).
+- `/plots/supplementary_plots` : comprehensive set of QC plots with greater detail, some of which are used as panels
+ in `main_plots`.
+- `/data` : various datafiles from the QC process such as family structures, a list of the samples
+ used for plotting, variant statistics, and lists of overlapping variants against any external call sets provided.
+
+### Plots description
+
+This section summarizes the most important plots used for overall call set QC found in the `/plots/main_plots`
+directory.
+
+Example plots are provided from a high-quality dataset consisting of 161 samples from the 1000 Genomes Project aligned
+with DRAGEN v3.7.8. Note that QC metrics and distributions will vary among data sets, particularly as sample size
+increases, and these examples are intended to provide a simple baseline for acceptable call set quality. Please see
+the [Recommendations](#recommendations) section for prescribed quality criteria.
+
+:::note
+The following plots are of variants passing all filters (i.e. with the `FILTER` status set to `PASS`). This is the
+default behavior of the QC plots generated in [FilterGenotypes](./fg).
+
+When running `MainVcfQc` as a standalone workflow, users may set the
+[bcftools_preprocessing_options](#optional--bcftools_preprocessing_options) argument to limit plotted variants based on `FILTER` status.
+For example, to limit to `PASS` variants for a VCF generated from [FilterGenotypes](./fg) use:
+```
+"bcftools_preprocessing_options": "-i 'FILTER~\"PASS\""
+```
+:::
+
+#### SV per genome
+
+Shows per-genome SV count distributions broken down by SV type. The top row is a site-level analysis, and
+the bottom row provides counts of individual alleles. Distributions of genotype quality (`GQ`), SV size, and
+frequency are also shown.
+
+
+
+#### SV counts
+
+Summarizes total variant counts by type, frequency, and size.
+
+
+
+#### Genotype distributions
+
+Distributions of genotypes across the call set.
+
+The left panel plots carrier frequency against allele frequency and can be used to visualize genotyping bias. In this
+plot, the rolling mean should not lie on the lower or upper extremes (all het. or all hom.).
+
+The middle panel plots the Hardy-Weinberg (HW) distribution of the cohort and provides estimates for the proportions of
+variants in HW equilibrium (or nominally in HW equilibrium before a multiple testing correction). The right-hand
+plots stratify HW by SV size.
+
+:::note
+The horizontal line bisecting the vertical axis in the AF vs. carrier frequency plot was fixed in.
+[#625](https://github.com/broadinstitute/gatk-sv/pull/625)).
+:::
+
+
+
+#### Size distributions
+
+Contains plots of SV length stratified by class and frequency.
+
+
+
+#### Frequency distributions
+
+Contains plots of allele frequency stratified by class and size.
+
+
+
+####
-- **GatherBatchEvidence** Copy number variant calling using
- `cn.MOPS` and `GATK gCNV`; B-allele frequency (BAF) generation;
- call and evidence aggregation.
+Each module is implemented in the [Workflow Description Language (WDL)](https://openwdl.org). The Terra workspaces come
+pre-configured with default values for all required parameters and set up to run the pipeline for most use cases.
-- **ClusterBatch** Variant clustering
+The following sections supplement the Terra workspaces with documentation for each WDL, including an overview of its
+function, key input parameters, and outputs. Not all parameters are documented here, as some WDLs contain dozens of
+inputs. Descriptions of some common inputs can be found in the [Resource files](/docs/resources) and
+[Runtime attributes](/docs/runtime_attr) sections. Users are encouraged to refer to the WDL source code for additional
+clarification.
-- **GenerateBatchMetrics** Variant filtering metric generation
-
-- **FilterBatch** Variant filtering; outlier exclusion
-
-- **GenotypeBatch** Genotyping
-
-- **MakeCohortVcf** Cross-batch integration; complex variant resolution and re-genotyping; vcf cleanup
-
-- **Module 07 (in development)** Downstream filtering, including minGQ, batch effect check,
- outlier samples removal and final recalibration;
-
-- **AnnotateVCF** Annotations, including functional annotation,
- allele frequency (AF) annotation and AF annotation with external population callsets;
-
-- **Module 09 (in development)** Visualization, including scripts that generates IGV screenshots and rd plots.
-
-- Additional modules to be added: de novo and mosaic scripts
-
-
-## Pipeline Parameters
-
-Several inputs are shared across different modules of the pipeline, which are explained in this section.
-
-#### `ped_file`
-
-A pedigree file describing the familial relationships between the samples in the cohort.
-The file needs to be in the
-[PED format](https://gatk.broadinstitute.org/hc/en-us/articles/360035531972-PED-Pedigree-format).
-Updated with [EvidenceQC](./eqc) sex assignments, including
-`sex = 0` for sex aneuploidies;
-genotypes on chrX and chrY for samples with `sex = 0` in the PED file will be set to
-`./.` and these samples will be excluded from sex-specific training steps.
+For details on running GATK-SV on Terra, refer to the [Execution](/docs/execution/joint#instructions) section.
diff --git a/website/docs/modules/join_raw_calls.md b/website/docs/modules/join_raw_calls.md
new file mode 100644
index 000000000..55b563a24
--- /dev/null
+++ b/website/docs/modules/join_raw_calls.md
@@ -0,0 +1,54 @@
+---
+title: JoinRawCalls
+description: Clusters unfiltered variants across batches
+sidebar_position: 17
+slug: jrc
+---
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/JoinRawCalls.wdl)
+
+This module clusters raw unfiltered variants from [ClusterBatch](./cb) across all batches. Concordance between these
+genotypes and the joint call set usually can be indicative of variant quality and is used downstream for genotype
+filtering.
+
+The following diagram illustrates the recommended invocation order:
+
+```mermaid
+
+stateDiagram
+ direction LR
+
+ classDef inModules stroke-width:0px,fill:#caf0f8,color:#00509d
+ classDef thisModule font-weight:bold,stroke-width:0px,fill:#ff9900,color:white
+ classDef outModules stroke-width:0px,fill:#caf0f8,color:#00509d
+
+ cb: ClusterBatch
+ jrc: JoinRawCalls
+ svc: SVConcordance
+ cb --> jrc
+ jrc --> svc
+
+ class jrc thisModule
+ class cb inModules
+ class svc outModules
+```
+
+### Inputs
+
+#### `prefix`
+Prefix for the output VCF, such as the cohort name. The guidelines outlined in the
+[sample ID requirements](/docs/gs/inputs#sampleids) section apply here.
+
+#### `clustered_*_vcfs`
+Array of clustered VCFs for all batches, generated in [ClusterBatch](./cb#clustered__vcf).
+
+#### `ped_file`
+Family structures and sex assignments determined in [EvidenceQC](./eqc). See [PED file format](/docs/gs/inputs#ped-format).
+
+### Outputs
+
+#### `joined_raw_calls_vcf`
+VCF containing all raw calls in the cohort.
+
+#### `ploidy_table`
+TSV of contig ploidies for all samples, assuming diploid autosome and sex assignments from the [ped_file](#ped_file).
diff --git a/website/docs/modules/main_vcf_qc.md b/website/docs/modules/main_vcf_qc.md
new file mode 100644
index 000000000..d362765b4
--- /dev/null
+++ b/website/docs/modules/main_vcf_qc.md
@@ -0,0 +1,228 @@
+---
+title: MainVcfQC
+description: VCF QC plotting
+sidebar_position: 21
+slug: mvqc
+---
+
+import { Highlight, HighlightOptionalArg } from "../../src/components/highlight.js"
+
+[WDL source code](https://github.com/broadinstitute/gatk-sv/blob/main/wdl/MainVcfQC.wdl)
+
+Creates plots for call set analysis and quality control (QC). This module is run by default with the
+[FilterGenotypes](./fg) module. Note, however, that the module is a stand-alone workflow and can be used with any batch-
+or cohort-level (i.e. multi-sample) SV VCF, including outputs from [ClusterBatch](./cb) and onward.
+
+Users can optionally provide external benchmarking datasets with the
+[site_level_comparison_datasets](#optional-site_level_comparison_datasets) and
+[sample_level_comparison_datasets](#optional-sample_level_comparison_datasets) parameters.
+
+The following sections provide guidance on interpreting these plots for VCFs that have been run with the recommended
+pipeline through [FilterGenotypes](./fg).
+
+### Output structure
+
+The tarball output has the following directory structure:
+
+- `/plots/main_plots` : main QC plots, described in detail [below](#plots-description).
+- `/plots/supplementary_plots` : comprehensive set of QC plots with greater detail, some of which are used as panels
+ in `main_plots`.
+- `/data` : various datafiles from the QC process such as family structures, a list of the samples
+ used for plotting, variant statistics, and lists of overlapping variants against any external call sets provided.
+
+### Plots description
+
+This section summarizes the most important plots used for overall call set QC found in the `/plots/main_plots`
+directory.
+
+Example plots are provided from a high-quality dataset consisting of 161 samples from the 1000 Genomes Project aligned
+with DRAGEN v3.7.8. Note that QC metrics and distributions will vary among data sets, particularly as sample size
+increases, and these examples are intended to provide a simple baseline for acceptable call set quality. Please see
+the [Recommendations](#recommendations) section for prescribed quality criteria.
+
+:::note
+The following plots are of variants passing all filters (i.e. with the `FILTER` status set to `PASS`). This is the
+default behavior of the QC plots generated in [FilterGenotypes](./fg).
+
+When running `MainVcfQc` as a standalone workflow, users may set the
+[bcftools_preprocessing_options](#optional--bcftools_preprocessing_options) argument to limit plotted variants based on `FILTER` status.
+For example, to limit to `PASS` variants for a VCF generated from [FilterGenotypes](./fg) use:
+```
+"bcftools_preprocessing_options": "-i 'FILTER~\"PASS\""
+```
+:::
+
+#### SV per genome
+
+Shows per-genome SV count distributions broken down by SV type. The top row is a site-level analysis, and
+the bottom row provides counts of individual alleles. Distributions of genotype quality (`GQ`), SV size, and
+frequency are also shown.
+
+
+
+#### SV counts
+
+Summarizes total variant counts by type, frequency, and size.
+
+
+
+#### Genotype distributions
+
+Distributions of genotypes across the call set.
+
+The left panel plots carrier frequency against allele frequency and can be used to visualize genotyping bias. In this
+plot, the rolling mean should not lie on the lower or upper extremes (all het. or all hom.).
+
+The middle panel plots the Hardy-Weinberg (HW) distribution of the cohort and provides estimates for the proportions of
+variants in HW equilibrium (or nominally in HW equilibrium before a multiple testing correction). The right-hand
+plots stratify HW by SV size.
+
+:::note
+The horizontal line bisecting the vertical axis in the AF vs. carrier frequency plot was fixed in.
+[#625](https://github.com/broadinstitute/gatk-sv/pull/625)).
+:::
+
+
+
+#### Size distributions
+
+Contains plots of SV length stratified by class and frequency.
+
+
+
+#### Frequency distributions
+
+Contains plots of allele frequency stratified by class and size.
+
+
+
+####