This is a repository for content relating to the Hackathon organised by EuroCC Cyprus with the Anticancer Society of Cyprus.
- Make sure your key setup is correct
- Login on cyclone
> ssh hack24gp##@cyclone.hpcf.cyi.ac.cy > sbatch hackathon/launch_hackathon.sub- Run command ssh command shown in connection_info.txt
- Connect to jupyter server shown in connection_info.txt
- Code your solution
- An excel of patient files. This can be found on the cluster under the hackathon folder in your account's home directory.
- You'll be given a small script which reads this into a Pandas DataFrame
- For acronyms in the frequency and route fields, see first sheet of hackathon.xlsx
- A list of pdf files containing the leaflets of medicines that are in the patient files.
Your submission should be a compressed zip or tar file containing your script and the json file described below. You can then send that compressed file by replying to the email Kyriaki sent to you with the team number. If you have used an external dataset, a small explanation in that email for how we can download it should be fine or even better have that as part of your script.
This should be submited by Sunday 13/10/2024 at 23:59!
You are expected to only list major interactions but minor ones will earn some extra points.
Major interactions: These are interactions which the Anticancer Society has given us to test your solutions. These will give you the majority of points.
Minor interactions: These are interactions which you may find in the medicine leaflets stating something along the lines of "speak with your doctor if you are taking Y medicine along with X". These should not be marked as a major interaction.
Bonus points for explained interactions. (e.g The two sedatives Oxycodone and Morphine shouldn't be consumed together because of their additive effects).
The format of your submission we expect looks like this (json):
{ "PATIENT 2": [
{ "interaction":["tramadol","pregabalin"],
"severity":"Major",
"explanation":"Using narcotic pain medications together with other medications that cause central nervous system depression can lead to respiratory distress, coma, and even death."
},
{ "interaction":["X","Y"],
"severity":"Minor",
"explanation":"There were no major interactions found in the dataset, however for medicine X it is suggested to consult with your doctor if taking X and Y together.
}
]
"PATIENT 3": [...]
}
In the case above the major interaction was correctly detected and explained, the next one is a minor interaction which will act as a warning.
If you try to check for all combination in a patient file, you'll need to check for a huge number of combinations.
Specifically 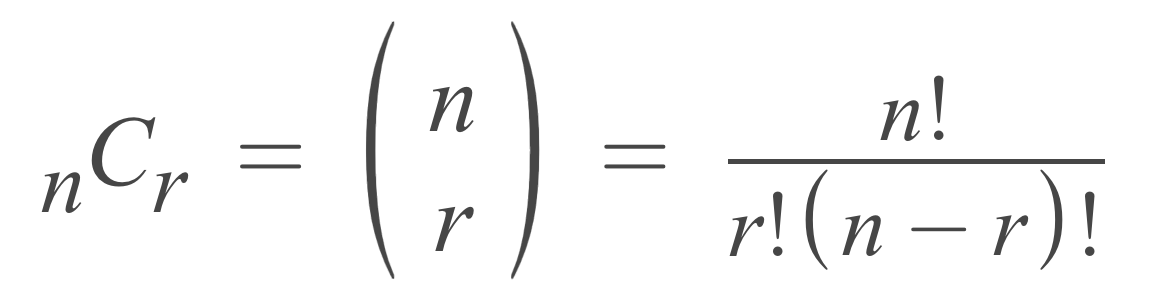
We prefer a script that will read through the the excel spreadsheet we provided and generate the answers in the format above, saving it to as a json file.
Winner will be announced in the near future when we evaluate your results.
Langchain: Useful for creating LLM prompt chains
Langgraph: Useful for debugging your chains
Langsmith: Useful for creating complex graph prompts
You will be limited to only use these models:
- gemma2:27b (Google)
- llama3.1:latest (Meta)
- phi3.5:3.8b-mini-instruct-fp16 (Microsoft)
- nemotron-mini:4b-instruct-fp16 (Nvidia)
be considerate of your context length as to not overflow to CPU memory
To help with your generation, read through the parameters you can set for ChatOllama
1 GPU - 32GB V100 from NVIDIA
10 CPU cores
40 GB RAM
https://www.medscape.com/pharmacists
https://go.drugbank.com/drug-interaction-checker#results
if ssh-agent is stopped (powershell):
Get-Service -Name ssh-agent | Set-Service -StartupType Manual
Start-Service ssh-agent
for permission issues (powershell):
icacls <path-to-your-id_rsa> /inheritance:r /grant:r "$($env:USERNAME):(F)"